Abstract
With the rapid evolution of diagnostic tools, particularly next-generation sequencing, the identification of genetic diseases, predominantly those with pediatric-onset, has significantly advanced. However, this progress presents challenges that span from selecting appropriate tests to the final interpretation of results. This review examines various genetic testing methodologies, each with specific indications and characteristics, emphasizing the importance of selecting the appropriate genetic test in clinical practice, taking into account factors like detection range, cost, turnaround time, and specificity of the clinical diagnosis. Interpretation of variants has become more challenging, often requiring further validation and significant resource allocation. Laboratories primarily classify variants based on the American College of Medical Genetics and Genomics and the Association for Clinical Genomic Science guidelines, however, this process has limitations. This review underscores the critical role of clinicians in matching patient phenotypes with reported genes/variants and considering additional factors such as variable expressivity, disease pleiotropy, and incomplete penetrance. These considerations should be aligned with specific gene-disease characteristics and segregation results based on an extended pedigree. In conclusion, this review aims to enhance understanding of the complexities of clinical genetic testing, advocating for a multidisciplinary approach to ensure accurate diagnosis and effective management of rare genetic diseases.
The landscape of diagnostic methodologies for genetic diseases has undergone a remarkable transformation, leading to the discovery of thousands of genetic conditions. Currently, around 7,000 rare diseases have been identified, with an estimated 80% attributed to genetic factors, and 50% manifesting during childhood [1-4]. Early genetic diagnosis has proven particularly beneficial for pediatric patients, offering significant cost savings and enabling long-term disease management [5]. Recent technological advancements and cost reductions have made various genetic tests integral to diagnostic evaluations in clinical practice. Next-generation sequencing (NGS) stands out due to its ability to simultaneously identify variants across multiple genes. This capability has led to cost efficiency and high diagnostic rates, especially for diseases characterized by genetic heterogeneity. Nevertheless, the challenge of interpreting the vast array of variants generated by NGS is non-trivial and often requires additional resources for validation. NGS may not be the optimal choice for single-gene diseases distinguishable by characteristic clinical features, and it is not considered the gold standard for detecting certain genetic variations such as copy number variations (CNVs) or short tandem repeats (STRs). Therefore, a critical aspect of the diagnostic process is accurate clinical assessment, followed by the judicious choice of testing methodology and careful interpretation of test reports by clinicians.
This review is divided into two sections. The first section covers various clinical genetic testing methodologies, discussing the types of tests and factors influencing test selection. The second section focuses on the interpretation of germline sequence variants, a common outcome of NGS. This structure allows for a comprehensive exploration of clinical genetic testing, ranging from broad methodologies to the detailed interpretation of specific variants.
Clinical genetic testing has become a pivotal component in diagnosing rare diseases, offering a broad range of tests available in clinical settings. Each genetic test is performed based on specific principles and has corresponding indications. There are three primary types of genetic testing: cytogenetic (chromosomal), DNA (molecular), and biochemical. Chromosomes, thread-like structures made up of DNA can be observed under a microscope after specific staining since the 1980s [6]. Microarray techniques, designed to identify small-unbalanced rearrangements, had developed and now feature diverse established platforms [7-9]. This method is recognized as a first-tier cytogenetic test for patients with developmental delay/intellectual disabilities, multiple congenital anomalies, and autism spectrum disorder [10-13]. Fluorescence in situ hybridization provides a unique advantage by visually mapping genetic material within a cell, facilitating the identification of structural chromosomal abnormalities [14,15]. Molecular genetic testing, the most frequently performed category, assesses single DNA loci, single genes, or multiple genes. Sanger sequencing, a traditional method and the gold standard for identifying single nucleotide variations is renowned for its high accuracy in analyzing short DNA sequences [16]. The polymerase chain reaction (PCR), a versatile tool widely used to amplify small DNA segments, is essential in various genetic tests, including those for infectious diseases [17-19]. Multiplex ligation-dependent probe amplification (MLPA), a robust method for detecting deletions and duplications of up to 50 nucleic acid sequences, proves invaluable for diagnosing various genetic disorders [20,21]. Some biochemical genetic tests, such as Southern blotting, while less common, still play a role in identifying specific DNA sequences in larger DNA samples [22,23]. NGS, a revolutionary form of molecular genetic testing, enables the rapid sequencing of large stretches of DNA or RNA, dramatically transforming the fields of genomics and molecular biology and facilitating a broad range of applications [24-27].
In clinical practice, each genetic testing method is tailored to a specific indication. In terms of the detection range, some tests focus on single loci, while others, like NGS, can cover the entire genome. However, NGS is not the gold standard for detecting CNVs or STRs. Despite a decrease in cost and processing time, NGS remains more expensive and has a longer turnaround time (TAT) compared to traditional tests. Clinicians must take these factors into account when selecting tests (Table 1). An accurate and specific clinical diagnosis is crucial, as it guides the identification of potential causative genes and common types of genetic variation. For example, in cases clinically diagnosed with Fragile X syndrome, the first-tier confirmatory tests are Southern blotting or PCR targeting FMR1. Similarly, for pathologically confirmed Alport syndrome with a family history of X-linked inheritance, testing for the COL4A5 gene using sequencing and MLPA is advisable, as 10% to 15% of these cases involve exon-level deletions or duplications [28,29]. A precise clinical diagnosis facilitates targeted testing, thereby reducing both the length of the diagnostic journey and associated costs. Although essential, a detailed clinical assessment, including examination findings, routine laboratory tests, and specific biomarkers such as pathological findings, is not extensively discussed in this article.
The urgency of diagnosis is vital in certain cases, necessitating consideration of clinical severity and TAT. Many inherited metabolic disorders can lead to irreversible damage if not promptly managed [30]. In cases of serious and rapidly progressive illnesses, quick decision-making is essential. Some patients may find themselves in a situation where they have a serious and rapidly progressive illness, requiring swift decision-making. In such cases, opting for tests with the fastest available results rather than the most cost-effective sequence of tests may be clinically justified. Rapid genomic sequencing, which significantly shortens TAT, is increasingly used for patients with suspected medically actionable disorders or those in intensive care units [31-33].
Finally, cost-effectiveness is a critical factor in healthcare, particularly for the diagnosis of rare diseases. The introduction of advanced genomic technologies like NGS has broadened our diagnostic scope, but their higher initial costs necessitate careful test selection. Cost-effectiveness typically involves starting with less expensive tests, followed by more comprehensive and sensitive yet costlier methods if the initial results are inconclusive. This tiered approach balances the need for thorough genetic analysis with budget constraints and enhances patient care by providing efficient and economically viable genetic testing strategies. Economic considerations in genetic testing go beyond cost reduction; they focus on maximizing the value that each test brings in terms of clinical outcomes and informed healthcare decisions. Although recent studies have demonstrated the cost-effectiveness of genome sequencing as a primary diagnostic tool in certain rare disease groups [34-36], the heterogeneity in study inclusions and cost-effectiveness parameters cast doubt on the generalizability of these results, highlighting the need for further well-designed studies. Additionally, costs and availability differ by country; for example, in Korea, the current insurance system officially covers only limited multi-gene panel tests, necessitating a different approach to the diagnostic strategy.
Various genetic variants contribute to the development of rare genetic disorders. However, this section focuses specifically on germline sequence variants, which have become increasingly significant due to the rise in NGS utilization. This technology generates numerous variants of uncertain clinical significance. The process of identifying variants through NGS data involves the following sequential steps: variant calling, annotation, and the evaluation of disease causality. In this section, we will delve into the process of evaluating the final disease causality of a variant as reported in the test, particularly from the clinician’s perspective.
Classifying germline sequence variants is a crucial step in genetic testing, guided by comprehensive criteria established by leading organizations such as the American College of Medical Genetics and Genomics (ACMG) and the Association for Clinical Genomic Science (ACGS). The widely recognized and utilized ACMG guideline categorizes variants into five groups: pathogenic, likely pathogenic, uncertain significance, likely benign, and benign. This classification relies on various factors, including the population database, gene characteristics, prior reports, segregation results, computational predictions, and functional studies [37]. ACGS provides a framework aligned with ACMG, underscoring the significance of clinical context and multidisciplinary expert consensus in variant interpretation [38]. Typically, laboratory physicians report pathogenic and likely pathogenic variants and occasionally variants of uncertain significance based on their policies. Despite global application, these guidelines are not without limitations. Variability in interpretation among clinicians and laboratories may result in inconsistent variant classification [39]. The databases crucial for variant interpretation are still evolving and may not sufficiently cover population-specific variations. Furthermore, these guidelines often prioritize molecular characteristics over the complete clinical profile of the patient, potentially leading to less personalized assessments. There is a growing demand for clearer variant interpretation, prompting efforts to modify and update these guidelines to better suit specific genetic or clinical subgroups [40-44]. However, studies based on large-scale cohorts have yet to provide universally applicable criteria for variant interpretation. Consequently, the final interpretation of variants, particularly those of uncertain significance, remains challenging when relying solely on guidelines. Achieving accurate interpretation and application of these findings necessitates a comprehensive, patient-centric approach by clinicians [45-48].
Before finalizing the results for a patient, clinicians must consider various factors in alignment with the reported genes/variants. The first factor is key phenotypes which include the age of onset, primary clinical symptoms, and disease progression. While theoretically, patients with the same variant may exhibit similar clinical symptoms, it is common to find individuals with the same variant presenting a wide range of diverse phenotypes, even within the same family (Fig. 1). This necessitates careful consideration [49]. For instance, the autosomal dominant polycystic kidney disease, is well known for various phenotypes, ranging from simple cyst to early end-stage renal disease, illustrating variable expressivity (Fig. 1A) [50]. The GLB1 gene, known to cause GM1-gangliosidosis with symptoms including progressive cerebral degeneration and developmental regression, is also responsible for Morquio disease. Morquio disease is characterized by multiple skeletal abnormalities and coarse facial features without clear neurological symptoms, exemplifying phenotypic variation or pleiotropy (Fig. 1B) [51,52]. A detailed family history assessment is crucial for patients with genetic disorders. The inheritance patterns of the identified genes should align with the family history and segregation results. If a gene associated with an autosomal dominant Mendelian disorder is documented, the variant should not be present in the asymptomatic parents, indicating de novo variation. Family test results, reflecting inheritance patterns, are critical and are incorporated into the ACMG and ACGS guidelines [37,38]. Unmatched results necessitate a reevaluation of the diagnosis. Notably, some diseases exhibit incomplete or reduced penetrance, explaining why asymptomatic family members carry the causative variant (Fig. 1D) [53,54]. Factors influencing penetrance include variant types, gene expression levels, epigenetic changes, gene-environment interactions, and genetic modifiers [55]. However, these theoretical factors often provide limited practical insight in clinical settings. Clinicians primarily rely on previous clinical reports and databases for practical information. For example, consider the case of the COL1A1 gene, associated with osteogenesis imperfecta, a rare connective tissue disorder. Osteogenesis imperfecta is known for its variable expression and incomplete penetrance, as documented in several clinical studies [56,57]. When a variant of the COL1A1 is identified in a family, and an asymptomatic family member carries the variant, it can be confirmed as causative if the phenotype matches the disease and co-segregation results for the rest of the family members (ideally from an extended pedigree) align with the known inheritance pattern, given the recognized incomplete penetrance of the gene. Similarly, specific diseases or gene subgroups, such as inherited retinal disease, hereditary spastic paraplegia, polycystic kidney disease, and renal agenesis/hypoplasia, are known to exhibit incomplete penetrance [58-61]. In summary, clinicians can ascertain the final causality of a variant using case-level clinical correlations. This determination hinges on a comprehensive assessment in which all pieces of the puzzle, including phenotypic consistency with the gene, family test results based on an extended pedigree, and research findings from existing databases, fit together harmoniously. If any of these factors are missing or inconclusive, a conservative interpretation approach should be adopted.
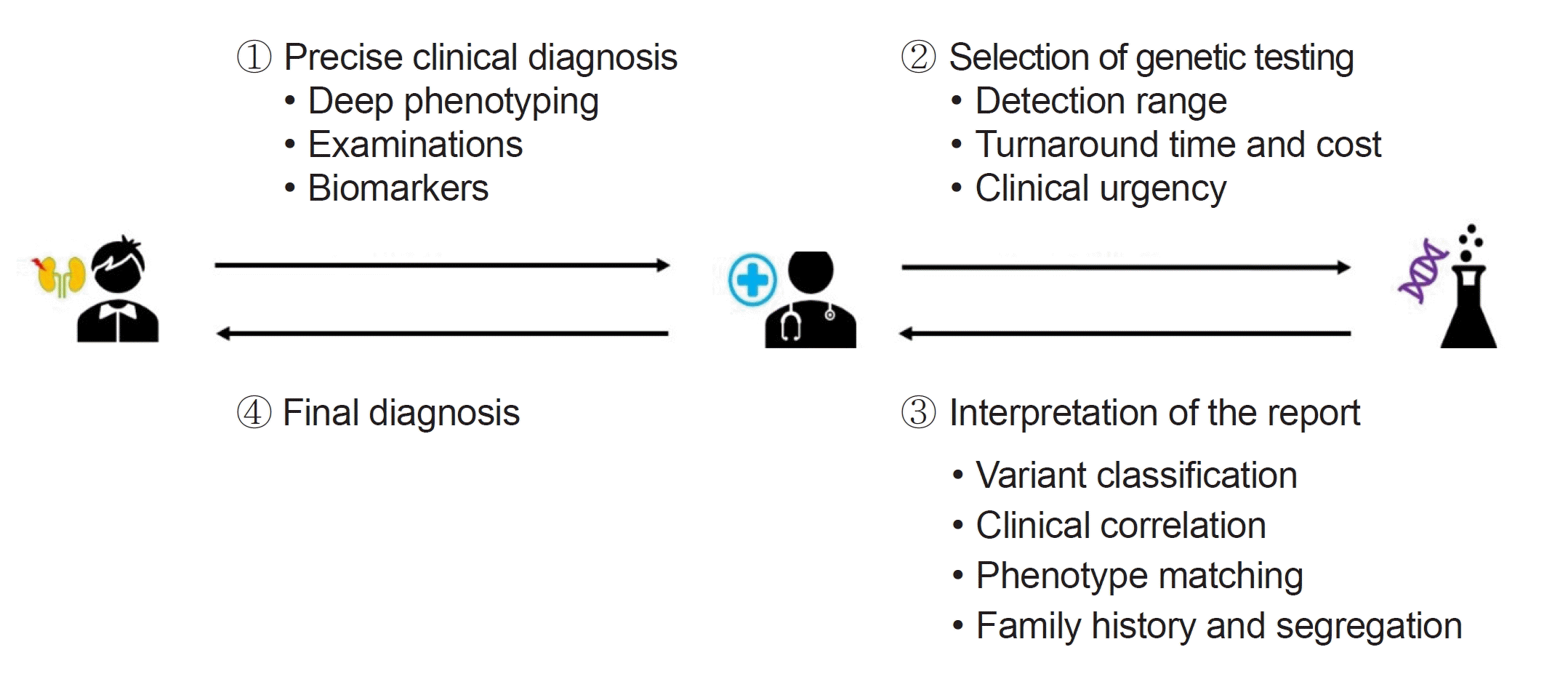
This review highlights the complexities and advancements in clinical genetic testing for rare diseases, emphasizing significant strides in diagnostic tool development, particularly NGS. While NGS has greatly improved our ability to identify genetic diseases, it has also necessitated meticulous validation. The selection of an appropriate genetic test in clinical practice requires careful consideration of factors such as detection range, cost, and clinical specificity. The role of germline variant classification, guided by the ACMG and ACGS guidelines, is crucial, but it faces limitations, including subjectivity and insufficient coverage of population-specific variations. To ensure accurate diagnosis and effective management, clinicians must meticulously align patient phenotypes with reported genes/variants, taking into consideration the variability in disease expression and extensive family histories (Fig. 2). This holistic and detailed approach is essential for enhancing patient care within the complex landscape of rare genetic diseases.
References
1. Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013; 14:681–91.

2. Orphanet activity report. Orphanet reports series/procedures [Internet]. Orphanet; 2022 [cited 2024 Jan 11]. Available from: https://www.orpha.net/consor/cgi-bin/Education_Home.php?lng=EN.
3. Nguengang Wakap S, Lambert DM, Olry A, Rodwell C, Gueydan C, Lanneau V, et al. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur J Hum Genet. 2020; 28:165–73.

4. Global Genes. RARE disease facts [Internet]. Global Genes; 2020 [cited 2024 Jan 11]. Available from: https://globalgenes.org/rare-facts/.
5. Lalonde E, Rentas S, Lin F, Dulik MC, Skraban CM, Spinner NB. Genomic diagnosis for pediatric disorders: revolution and evolution. Front Pediatr. 2020; 8:373.

6. Yunis JJ. Mid-prophase human chromosomes: the attainment of 2000 bands. Hum Genet. 1981; 56:293–8.

7. Kallioniemi OP, Kallioniemi A, Sudar D, Rutovitz D, Gray JW, Waldman F, et al. Comparative genomic hybridization: a rapid new method for detecting and mapping DNA amplification in tumors. Semin Cancer Biol. 1993; 4:41–6.
8. Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, et al. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997; 20:399–407.

9. Albertson DG, Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum Mol Genet. 2003; 12 Spec No 2:R145–52.

10. Ahn JW, Bint S, Bergbaum A, Mann K, Hall RP, Ogilvie CM. Array CGH as a first line diagnostic test in place of karyotyping for postnatal referrals: results from four years’ clinical application for over 8,700 patients. Mol Cytogenet. 2013; 6:16.

11. Battaglia A, Doccini V, Bernardini L, Novelli A, Loddo S, Capalbo A, et al. Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features. Eur J Paediatr Neurol. 2013; 17:589–99.

12. Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010; 61:437–55.

13. Vissers LE, de Vries BB, Veltman JA. Genomic microarrays in mental retardation: from copy number variation to gene, from research to diagnosis. J Med Genet. 2010; 47:289–97.

14. Rudkin GT, Stollar BD. High resolution detection of DNA-RNA hybrids in situ by indirect immunofluorescence. Nature. 1977; 265:472–3.

15. Iqbal MA, Ulmer C, Sakati N. Use of FISH technique in the diagnosis of chromosomal syndromes. East Mediterr Health J. 1999; 5:1218–24.

16. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977; 74:5463–7.

17. Goossens M. The amplification of nucleotide sequences by PCR and the new technics for molecular diagnosis. Reprod Nutr Dev. 1990; Suppl 1:117s–124s.
18. Ben-Ezra JM. Amplification methods in the molecular diagnosis of genetic diseases. Clin Lab Med. 1995; 15:795–815.

19. Erlich HA, Arnheim N. Genetic analysis using the polymerase chain reaction. Annu Rev Genet. 1992; 26:479–506.

20. Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002; 30:e57.

21. Stuppia L, Antonucci I, Palka G, Gatta V. Use of the MLPA assay in the molecular diagnosis of gene copy number alterations in human genetic diseases. Int J Mol Sci. 2012; 13:3245–76.

22. Borst M, Miller DM. DNA isolation and Southern analysis: a clinician's view. Am J Med Sci. 1990; 299:356–60.
23. Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975; 98:503–17.

24. Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015; 58:586–97.

25. Pettersson E, Lundeberg J, Ahmadian A. Generations of sequencing technologies. Genomics. 2009; 93:105–11.

26. Fernandez-Marmiesse A, Gouveia S, Couce ML. NGS technologies as a turning point in rare disease research, diagnosis and treatment. Curr Med Chem. 2018; 25:404–32.
27. Chung CC, Hue SP, Ng NY, Doong PH; Hong Kong Genome Project, Chu AT, et al. Meta-analysis of the diagnostic and clinical utility of exome and genome sequencing in pediatric and adult patients with rare diseases across diverse populations. Genet Med. 2023; 25:100896.

28. Jais JP, Knebelmann B, Giatras I, De Marchi M, Rizzoni G, Renieri A, et al. X-linked Alport syndrome: natural history and genotype-phenotype correlations in girls and women belonging to 195 families: a “European Community Alport Syndrome Concerted Action” study. J Am Soc Nephrol. 2003; 14:2603–10.
29. Bekheirnia MR, Reed B, Gregory MC, McFann K, Shamshirsaz AA, Masoumi A, et al. Genotype-phenotype correlation in X-linked Alport syndrome. J Am Soc Nephrol. 2010; 21:876–83.

30. Rao AN, Kavitha J, Koch M, Suresh Kumar V. Inborn errors of metabolism: review and data from a tertiary care center. Indian J Clin Biochem. 2009; 24:215–22.

31. Kim MJ, Kim SY, Lee JS, Kang S, Park LJ, Choi W, et al. Rapid targeted sequencing using dried blood spot samples for patients with suspected actionable genetic diseases. Ann Lab Med. 2023; 43:280–9.

32. Owen MJ, Niemi AK, Dimmock DP, Speziale M, Nespeca M, Chau KK, et al. Rapid sequencing-based diagnosis of thiamine metabolism dysfunction syndrome. N Engl J Med. 2021; 384:2159–61.

33. Wojcik MH, Callahan KP, Antoniou A, Del Rosario MC, Brunelli L, ElHassan NO, et al. Provision and availability of genomic medicine services in level IV neonatal intensive care units. Genet Med. 2023; 25:100926.

34. Incerti D, Xu XM, Chou JW, Gonzaludo N, Belmont JW, Schroeder BE. Cost-effectiveness of genome sequencing for diagnosing patients with undiagnosed rare genetic diseases. Genet Med. 2022; 24:109–18.

35. Runheim H, Pettersson M, Hammarsjo A, Nordgren A, Henriksson M, Lindstrand A, et al. The cost-effectiveness of whole genome sequencing in neurodevelopmental disorders. Sci Rep. 2023; 13:6904.

36. Yeung A, Tan NB, Tan TY, Stark Z, Brown N, Hunter MF, et al. A cost-effectiveness analysis of genomic sequencing in a prospective versus historical cohort of complex pediatric patients. Genet Med. 2020; 22:1986–93.

37. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–24.

38. Ellard S, Baple EL, Callaway A, Berry I, Forrester N, Turnbull C, et al. ACGS best practice guidelines for variant classification in Rare Disease 2020 [Internet]. Association for Clinical Genomic Science; 2020 [cited 2024 Jan 11]. Available from: https://www.acgs.uk.com/media/11631/uk-practice-guidelines-for-variant-classification-v4-01-2020.pdf.
39. Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am J Hum Genet. 2016; 98:1067–76.

40. Inoue Y, Machida O, Kita Y, Yamamoto T. Need for revision of the ACMG/AMP guidelines for interpretation of X-linked variants. Intractable Rare Dis Res. 2022; 11:120–4.

41. Patel MJ, DiStefano MT, Oza AM, Hughes MY, Wilcox EH, Hemphill SE, et al. Disease-specific ACMG/AMP guidelines improve sequence variant interpretation for hearing loss. Genet Med. 2021; 23:2208–12.

42. Strande NT, Brnich SE, Roman TS, Berg JS. Navigating the nuances of clinical sequence variant interpretation in Mendelian disease. Genet Med. 2018; 20:918–26.

43. Gelb BD, Cave H, Dillon MW, Gripp KW, Lee JA, Mason-Suares H, et al. ClinGen’s RASopathy Expert Panel consensus methods for variant interpretation. Genet Med. 2018; 20:1334–45.

44. Savige J, Storey H, Watson E, Hertz JM, Deltas C, Renieri A, et al. Consensus statement on standards and guidelines for the molecular diagnostics of Alport syndrome: refining the ACMG criteria. Eur J Hum Genet. 2021; 29:1186–97.

45. Chen E, Facio FM, Aradhya KW, Rojahn S, Hatchell KE, Aguilar S, et al. Rates and classification of variants of uncertain significance in hereditary disease genetic testing. JAMA Netw Open. 2023; 6:e2339571.

46. Burke W, Parens E, Chung WK, Berger SM, Appelbaum PS. The challenge of genetic variants of uncertain clinical significance : a narrative review. Ann Intern Med. 2022; 175:994–1000.

47. Johnson B, Ouyang K, Frank L, Truty R, Rojahn S, Morales A, et al. Systematic use of phenotype evidence in clinical genetic testing reduces the frequency of variants of uncertain significance. Am J Med Genet A. 2022; 188:2642–51.

48. Nykamp K, Anderson M, Powers M, Garcia J, Herrera B, Ho YY, et al. Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017; 19:1105–17.

49. Kingdom R, Wright CF. Incomplete penetrance and variable expressivity: from clinical studies to population cohorts. Front Genet. 2022; 13:920390.

50. Rossetti S, Kubly VJ, Consugar MB, Hopp K, Roy S, Horsley SW, et al. Incompletely penetrant PKD1 alleles suggest a role for gene dosage in cyst initiation in polycystic kidney disease. Kidney Int. 2009; 75:848–55.

51. Benson PF, Barbarik A, Brown SP, Mann TP. GM1-generalized gangliosidosis variant with cardiomegaly. Postgrad Med J. 1976; 52:159–65.

52. Arbisser AI, Donnelly KA, Scott CI Jr, DiFerrante N, Singh J, Stevenson RE, et al. Morquio-like syndrome with beta galactosidase deficiency and normal hexosamine sulfatase activity: mucopolysacchariodosis IVB. Am J Med Genet. 1977; 1:195–205.

53. Vytopil M, Ricci E, Dello Russo A, Hanisch F, Neudecker S, Zierz S, et al. Frequent low penetrance mutations in the Lamin A/C gene, causing Emery Dreifuss muscular dystrophy. Neuromuscul Disord. 2002; 12:958–63.

54. Burdon KP, Wirth MG, Mackey DA, Russell-Eggitt IM, Craig JE, Elder JE, et al. A novel mutation in the Connexin 46 gene causes autosomal dominant congenital cataract with incomplete penetrance. J Med Genet. 2004; 41:e106.

55. Shawky RM. Reduced penetrance in human inherited disease. Egypt J Medl Hum Genet. 2014; 15:103–11.

56. Pereira R, Halford K, Sokolov BP, Khillan JS, Prockop DJ. Phenotypic variability and incomplete penetrance of spontaneous fractures in an inbred strain of transgenic mice expressing a mutated collagen gene (COL1A1). J Clin Invest. 1994; 93:1765–9.

57. Van Dijk FS, Sillence DO. Osteogenesis imperfecta: clinical diagnosis, nomenclature and severity assessment. Am J Med Genet A. 2014; 164A:1470–81.

58. Ellingford JM, Hufnagel RB, Arno G. Phenotype and genotype correlations in inherited retinal diseases: population-guided variant interpretation, variable expressivity and incomplete penetrance. Genes (Basel). 2020; 11:1274.

59. Vujic M, Heyer CM, Ars E, Hopp K, Markoff A, Orndal C, et al. Incompletely penetrant PKD1 alleles mimic the renal manifestations of ARPKD. J Am Soc Nephrol. 2010; 21:1097–102.

Fig. 1.
Conceptual representation of expressivity, pleiotropy, and penetrance in autosomal dominant genetic disorders. (A) A pedigree displaying an autosomal dominant genetic with varying levels of disease expressivity among family members. (B) A pedigree illustrating an autosomal dominant genetic disorder demonstrating disease pleiotropy within family members. (C) A pedigree of autosomal dominant genetic disorder exhibiting complete penetrance. (D) A pedigree of autosomal dominant genetic disorder exhibiting incomplete penetrance. Mut, mutation.
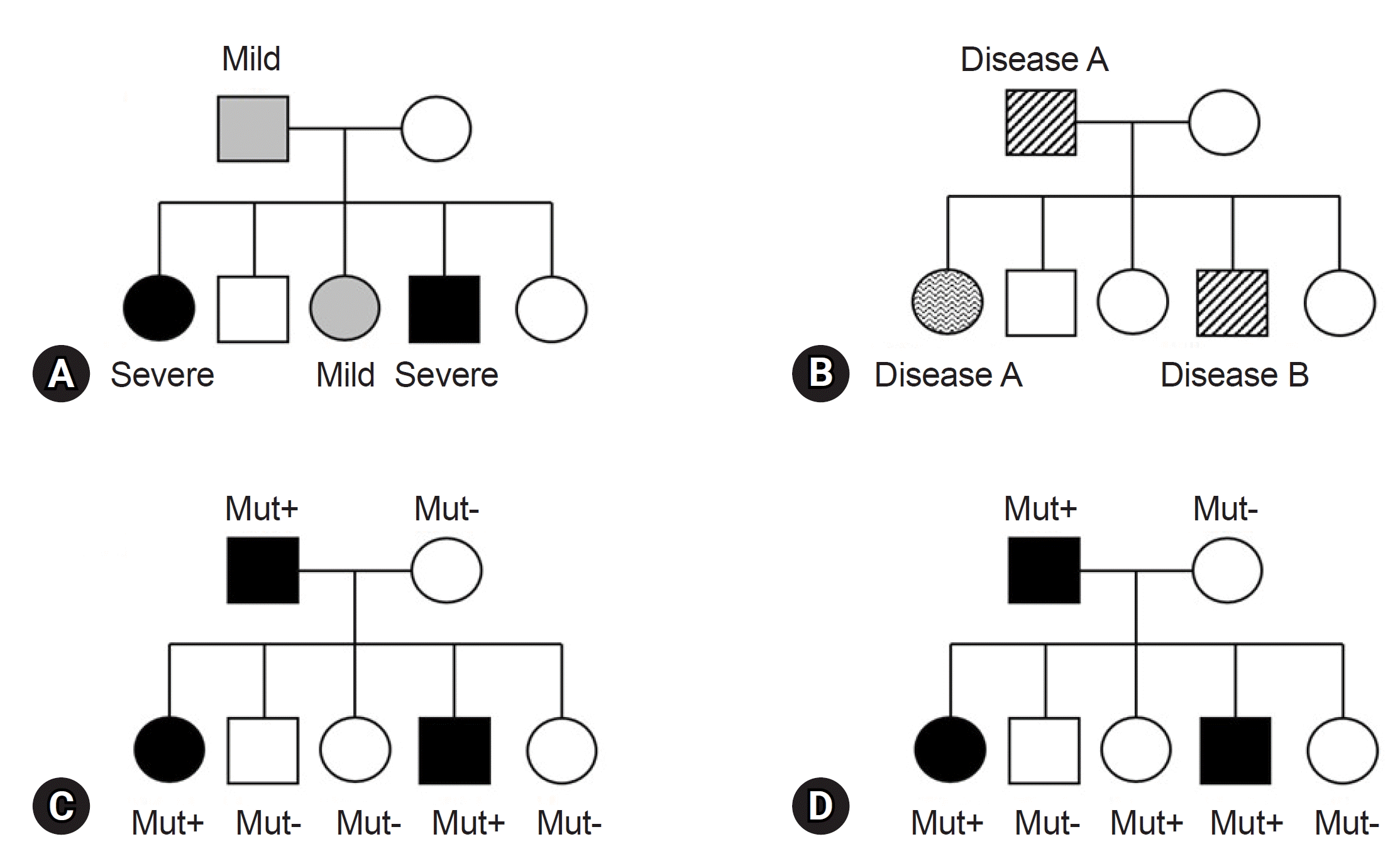
Table 1.
Comparative characteristics of different genetic testing methodologies




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download