Abstract
Solute carrier 40A1 (SLC40A1) encodes ferroportin, which is the only known transmembrane protein that exports elemental iron from mammalian cells and is essential for iron homeostasis. Mutations in SLC40A1 are associated with iron-overload disorders. In addition to ferroportin diseases, SLC40A1 expression is downregulated in various cancer types. Despite the clinical significance of the SLC40A1 transporter, only a few studies have investigated genetic variants in SLC40A1. The present study was performed to identify genetic variations in the SLC40A1 promoter and functionally characterize each variant using in vitro assays. We investigated four haplotypes and five variants in the SLC40A1 promoter. We observed that haplotype 3 (H3) had significantly lower promoter activity than H1, whereas the activity of H4 was significantly higher than that of H1. Luciferase activity of H2 was comparable to that of H1. In addition, four variants of SLC40A1, c.-1355G>C, c.-662C>T, c.-98G>C, and c.-8C>G, showed significantly increased luciferase activity compared to the wild type (WT), whereas c.-750G>A showed significantly decreased luciferase activity compared to the WT. Three transcription factors, cAMP response element-binding protein-1 (CREB-1), chicken ovalbumin upstream promoter transcription factor 1, and hepatic leukemia factor (HLF), were predicted to bind to the promoter regions of SLC40A1 near c.-662C>T, c.-98G>C, and c.-8C>G, respectively. Among these, CREB-1 and HLF bound more strongly to the variant sequences than to the WT and functioned as activators of SLC40A1 transcription. Collectively, our findings indicate that the two SLC40A1 promoter haplotypes affect SLC40A1 transcription, which is regulated by CREB-1 and HLF.
Solute carrier 40A1 (SLC40A1) encodes ferroportin, which exports iron from cells to the plasma. It is expressed in hepatocytes, macrophages, monocytes, and basolateral membranes of enterocytes. Hepcidin, produced by hepatocytes, binds to ferroportin and plays an important role in maintaining iron homeostasis by controlling its internalization and degradation [1]. Mutations in SLC40A1 are associated with iron-overload disorders. Two types of diseases are caused by loss-of-function or gain-of-function of this transporter. Loss-of-function of the transporter results in type A ferroportin disease, which is clinically characterized by normal or low transferrin saturation, hyperferritinemia, and iron accumulation. The gain-of-function of the transporter leads to type B ferroportin disease (also known as autosomal dominant type 4 hemochromatosis), and patients with this disease exhibit high transferrin saturation, hyperferritinemia, and iron overload [2,3]. For example, the missense mutant p.W158C causes defective trafficking of ferroportin to the plasma membrane, and one patient with this mutation showed normal transferrin saturation, hyperferritinemia, and iron overload in macrophages. In contrast, patients with another missense mutation, p.H506R, which causes resistance to hepcidin, have clinical features such as increased transferrin saturation, hyperferritinemia, and iron accumulation in hepatocytes [4].
In addition to ferroportin diseases, SLC40A1 is associated with several cancers. It has been reported that the expression of SLC40A1 is downregulated in various cancers, including breast, prostate, lung, and ovarian cancers, because iron plays an important role in numerous processes of carcinogenesis [5-8]. For example, Pinnix et al. [8] reported that the expression of SLC40A1 is reduced in breast cancer, and that this decreased expression correlates with the worst prognoses.
Despite the clinical importance of the SLC40A1 transporter, a few functional single nucleotide polymorphisms (SNPs) in SLC40A1 have been characterized. In this study, we investigated the effect of common haplotypes or genetic variants in the SLC40A1 promoter on its activity and the mechanism by which these SLC40A1 variants alter promoter activity.
SNP data were obtained from the database of SNP (dbSNP) of the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/snp/) to identify the common SNPs (minor allele frequency ≥ 5%) in the promoter region (within –1.5 kb from the translational start site) of SLC40A1. The frequencies of these variants were obtained using data from the 1,000 Genomes Project (Phase 3) for four ethnic groups: 661 African, 347 American, 504 East Asian, and 503 European individuals. Haplotype assembly was performed using the Haploview 4.3 software (Broad Institute). Nucleotide location numbers were assigned from the translational start site based on the SLC40A1 mRNA sequence (GenBank accession number: NM_014585.5).
A 1,554 bp section of the SLC40A1 gene was amplified using primers containing KpnI and HindIII recognition sites to construct a reporter plasmid containing SLC40A1 wild type (WT). Then, the amplified product was inserted into the pGL4.11 [luc2P] vector (Promega Corporation). The plasmids containing the SLC40A1 haplotypes or variants were generated using the QuikChange II site-directed mutagenesis kit (Agilent Technologies). All the primers used are listed in Supplementary Table 1. All the vector DNA sequences were confirmed by direct sequencing.
First, HCT-116 cells (human colon carcinoma) were seeded (1.2 × 105 cells/well) in a 24-well plate. The next day, 500 ng of reporter plasmids containing SLC40A1 WT or variants and 10 ng of pGL4.74 Renilla vector (Promega Corporation) were transfected into HCT-116 cells using Lipofectamine LTX and Plus reagents (Life Technologies). After 48 h, luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega Corporation), according to the manufacturer’s protocol. To examine effect of cAMP response element-binding protein-1 (CREB-1), chicken ovalbumin upstream promoter transcription factor 1 (COUP-TF1), and hepatic leukemia factor (HLF) on the promoter activity of SLC40A1, SLC40A1 reporter plasmids were co-transfected with increasing amounts (25–500 ng) of CREB-1 (Origene Technologies Inc.), COUP-TF1 (Origene Technologies Inc.), or HLF (Horizon Discovery) cDNA into HCT-116 cells. Relative luciferase activity was defined as the ratio of firefly luciferase to renilla luciferase using the pGL4.74 Renilla vector.
DNA-protein binding assays were performed using to 15 or 30 ug nuclear extract from HCT-116 cells. The interaction between nuclear extract and biotin-labeled (labeled at 5’) oligonucleotides was determined using DNA-protein binding assay kit (Colorimetric) (Abcam), according to the manufacturer’s protocol. Antibodies against CREB-1 (Abcam), COUP-TF1 (Santa Cruz Biotechnology), and HLF (Santa Cruz Biotechnology) were used in this assay. Absorbance was measured at 450 nm using a VERSAmax tunable microplate reader (Molecular Devices). The binding activity between the DNA and proteins was calculated according to the manufacturer's protocol. All oligonucleotides used in the DNA-protein binding assays are listed in Supplementary Table 1.
Statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software Inc.). p-values of the luciferase assays were calculated using a one-way analysis of variance (ANOVA), followed by Dunnett’s two-tailed test. p-values of the DNA-protein binding assays were calculated using the Student’s t-test. Statistical significance was set at p < 0.05.
Using SNP data from the NCBI dbSNP, we examined the common SNPs (minor allele frequency ≥ 5%) in the promoter region (within −1.5 kb from the translational start site) of SLC40A1 and identified the following eight SNPs: c.-1470C>T (rs16831699), c.-1461T>C (rs77227487), c.-1355G>C (rs3811621), c.-1098G>A (rs10202029), c.-750G>A (rs13015236), c.-662C>T (rs12693542), c.-98G>C (rs13008848), and c.-8C>G (rs11568351). The frequencies of these variants in four ethnic groups (661 African, 347 American, 504 East Asian, and 503 European individuals) were obtained from the 1000 Genomes Project (Phase 3) (Table 1). Table 2 shows the frequencies of major haplotypes SLC40A1 promoter region. Haplotype 3 (H3) contained the SLC40A1 WT mRNA sequence (NM_014585.5). H1 included three variants, c.-1355G>C, c.-750G>A, and c.-662C>T and H2 consisted of c.-1355G>C and c.-662C>T. H4 was composed of two variants, c.-98G>C and c.-8C>G. Among the haplotypes, H1 showed the highest frequencies in the three ethnic groups, East Asians, Americans, and Europeans. In Africans, H2 had the highest frequency (27.2%), and seven other haplotypes were found at frequencies of 5.5% to 9.9% (Supplementary Table 2).
To examine the effects of variants on SLC40A1 promoter activity, we constructed reporter plasmids containing SLC40A1 haplotypes or variants and performed luciferase assays to measure the luciferase activities of SLC40A1 promoter haplotypes, which were compared to that of H1, the most common haplotype in the East Asian, American, and European groups. H3 showed significantly decreased luciferase activity (26.7%), whereas H4 showed significantly increased promoter activity (40.2%) compared to H1. The promoter activity of H2 was comparable to that of the H1 (Fig. 1A). Next, we measured the luciferase activity of five SNPs: c.-1355G>C, c.-750G>A, c.-662C>T, c.-98G>C, and c.-8C>G in those four haplotypes. Their activities were then compared with those of the WT. The activities of four variants, c.-1355G>C, c.-662C>T, c.-98G>C, and c.-8C>G, were significantly increased compared to those of the WT. The activity of c.-750G>A was significantly decreased by 14.6% compared with that of the WT (Fig. 1B).
First, the transcription factors that affect the regulation of promoter activity for each SLC40A1 variant were predicted using Consite (http://consite.genereg.net). As a result, three transcription factors, CREB-1, COUP-TF1, and HLF, that can bind to the promoter region of SLC40A1 near c.-662C>T, c.-98G>C, and c.-8C>G and there was quite difference in the binding affinity of each transfection factor between the WT and variant sequences. To validate this prediction, we conducted a DNA-protein binding assay. As a result, CREB-1 more strongly bound to the c.-662T variant sequences by 15.1% than c.-662C WT sequences (Fig. 2A). In addition, we observed that COUP-TF1 bound 19.2% more strongly to the c.-98C variant sequences than to the c.-98G WT sequences (Fig. 2B). Finally, we found that HLF bound more strongly to the c.-8G variant sequences by 36.9% than c.-8C WT sequences (Fig. 2C).
Luciferase assays were performed after the co-transfection of SLC40A1 WT or variants and each transcription factor into HCT-116 cells to examine the effects of CREB-1, COUP-TF1, and HLF on SLC40A1 promoter activity (Fig. 3). CREB-1 and HLF significantly increased the luciferase activity (Fig. 3A, C), whereas COUP-TF1 decreased the luciferase activity in a dose-dependent manner (Fig. 3B). To determine whether overexpression of transcription factors affects cell viability, we performed cell viability assays using cell counting kit-8 assay kit (Dojindo Laboratories Co.) after co-transfecting HCT-116 cells with reporter plasmids containing SLC40A1 WT or variants and transcription factors—CREB-1, COUP-TF1, and HLF. The amount of each transcription factor was selected based on the largest amount used in the luciferase assay. We compared whether cell viability differed when the transcription factors were overexpressed compared to when they were not overexpressed. We found that none of the transcription factors had any significant effect on the cell viability (Supplementary Fig. 1). In addition, using a luciferase assay after co-transfection of a SLC40A1 WT reporter plasmid and five transcription factors—GATA binding protein 2 (GATA-2), nuclear factor erythroid 2 (NF-E2), nuclear transcription factor Y (NF-Y), NK2 transcription factor related, locus 5 (NKX-2.5), and sex-determining region Y gene (SRY)—that were predicted not to bind to the SLC40A1 promoter, we confirmed that these transcription factors had no significant effect on the SLC40A1 promoter activity (Supplementary Fig. 2).
In the present study, we investigated the effects of SLC40A1 promoter variants on its promoter activity. First, we measured the luciferase activity of the four major promoter haplotypes and observed that two changed the activity significantly compared to that of H1. H3 showed significantly decreased luciferase activity, whereas H4 showed significantly increased luciferase activity. We then measured the luciferase activity of five variants and observed that four promoter variants, c.-1355G>C, c.-662C>T, c.-98G>C, and c.-8C>G, significantly increased luciferase activity, whereas one variant, c.-750G>A, showed significantly decreased luciferase activity.
Three transcription factors, CREB-1, COUP-TF1, and HLF, were predicted to bind near the variants c.-662C>T, c.-98G>C, and c.-8C>G, respectively, and there was a large difference in the binding affinity of each transcription factor between the WT and variant sequences. The results of the DNA-protein binding assay were consistent with this prediction. We found that all three transcription factors bound to the variant sequences were stronger than those bound to the WT sequences.
CREB-1 is expressed in various cell types and regulates promoter activity by binding to cAMP-responsive elements [9]. Previous studies have shown various role for CREB-1 in the body, such as in B cell development, memory performance, and promotion of hepatic fibrosis [10-12]. In addition, CREB-1 is related with poor prognosis in breast cancer and increased susceptibility to major psychiatric disorders [13,14]. Nabokina et al. [15] reported that CREB-1 binds to the promoter of SLC44A4, a human thiamine pyrophosphate transporter, and activates its promoter.
COUP-TF1 plays important roles in normal brain development, regulation of hepatitis B virus transcription, and differentiation of oligodendrocytes [16-18]. COUP-TF1 is associated with several cancers. For example, COUP-TF1 acts as a barrier to the dissemination of early-stage breast cancer cells [19]. Another study reported that the expression of COUP-TF1 is inversely correlated with androgen levels, which play an important role in prostate cancer [20].
HLF (originally known as E2A-HLF) is a member of the PAR subfamily of basic leucine-zipper proteins [21]. HLF regulates hematopoietic cell development, and ectopic expression of HLF promotes cell death [22,23]. In addition, HLF is involved in the development of hepatocellular carcinoma and liver fibrosis [24,25]. In childhood acute lymphocytic leukemia, E2A-HLF generated by the translocation [17,19] is associated with a high mortality rate [26]. Interestingly, in the case of gliomas, HLF is downregulated compared to that in the normal brain and is related to a higher survival rate [27]. Recently, we reported that HLF binds to the ATP-binding cassette subfamily 4 (ABCA4) promoter and functions as an activator of ABCA4 transcription [28].
In the present study, we observed that CREB-1 and HLF activated the transcription of SLC40A1, whereas COUP-TF1 functioned as a repressor. These data suggest that the increased luciferase activities of the variants c.-662T and c.-8G are related to stronger binding affinities of CREB-1 and HLF, respectively, to variant sequences than to WT sequences. In the case of c.-98G>C, the variant c.-98C showed significantly increased luciferase activity, although the binding affinity of COUP-TF1 was stronger for this variant than for the WT. This finding indicated that transcription factors other than COUP-TF1 are involved in the increased transcriptional activity of c.-98G>C.
In most cases, genetic testing for personalized drug treatment is performed when the drug target is a gene [29]. However, researchers continue to report genetic research results and present clinical guidelines to provide personalized treatment to patients based on their genetic information. For example, the guideline based on solute carrier organic anion transporter family member 1B1 (SLCO1B1), ATP-binding cassette G2 (ABCG2), and cytochrome P450 2C9 (CYP2C9) genotype information was recently reported to improve the safety and efficacy of statins [30]. This study revealed that two SLC40A1 promoter haplotypes significantly change promoter activity in vitro. In addition to missense mutants that cause gain- or loss-of-function of the transporter, genetic variants within the promoter, such as those in this study, can have a significant impact on disease susceptibility or course. For example, haplotypes consisting of the promoter variant of organic cation/carnitine transporter 2 (OCTN2), c.-207G>C, and the p.L503F variant of OCTN1 are significantly correlated with susceptibility to Crohn's disease [31-34]. We previously reported that OCTN1 or OCTN2 promoter haplotypes that significantly reduce the activity of their promoters have a significant impact on the progression of Crohn's disease in Koreans [35,36]. Further studies will be needed to determine whether SLC40A1 promoter haplotypes have clinical utility for diseases related to the SLC40A1 transporter.
SLC40A1 transporter functions as an exporter of iron outside the cells. The loss-of-function or gain-of-function of this transporter caused by missense mutants results in iron-overload disorders [2,3]. The expression of SLC40A1 is associated with various cancers. For example, the accumulation of iron ions in the body due to decreased expression of SLC40A1 is significantly correlated with an increased risk and progression of colon cancer [37]. Recently, it was found that inhibiting the expression of SLC40A1 promotes the proliferation of prostate cancer [38]. In addition, the expression of SLC40A1 is known to affect the response to anticancer treatment in patients with cancer. Previously it has been reported that inhibition of SLC40A1 expression induces cisplatin resistance in ovarian cancer [39]. Another study found that reduced SLC40A1 expression increases sensitivity to chemotherapy in patients with acute myeloid leukemia [40].
In the present study, we found that two SLC40A1 haplotypes changed their promoter activity, and that CREB-1 and HLF were involved in transcriptional regulation. To the best of our knowledge, this is the first study to examine the effect of genetic variants in the SLC40A1 on its promoter. Further studies are needed to determine whether these haplotypes affect iron transport in the body or whether they affect the susceptibility and prognosis of other disorders such as cancer.
Supplementary data including two tables and two figures can be found with this article online at https://doi.org/10.4196/kjpp.2024.28.2.113
Notes
REFERENCES
1. Ravasi G, Pelucchi S, Russo A, Mariani R, Piperno A. 2020; Ferroportin disease: a novel SLC40A1 mutation. Dig Liver Dis. 52:688–690. DOI: 10.1016/j.dld.2020.03.013. PMID: 32360131.

2. Moreno-Carralero MI, Muñoz-Muñoz JA, Cuadrado-Grande N, López-Rodríguez R, José Hernández-Alfaro M, Enríquez-de-Salamanca R, Méndez M, Morán-Jiménez MJ. del-Castillo-Rueda A. 2014; A novel mutation in the SLC40A1 gene associated with reduced iron export in vitro. Am J Hematol. 89:689–694. DOI: 10.1002/ajh.23714. PMID: 24644245.
3. Détivaud L, Island ML, Jouanolle AM, Ropert M, Bardou-Jacquet E, Le Lan C, Mosser A, Leroyer P, Deugnier Y, David V, Brissot P, Loréal O. 2013; Ferroportin diseases: functional studies, a link between genetic and clinical phenotype. Hum Mutat. 34:1529–1536. DOI: 10.1002/humu.22396. PMID: 23943237.

4. Mayr R, Griffiths WJ, Hermann M, McFarlane I, Halsall DJ, Finkenstedt A, Douds A, Davies SE, Janecke AR, Vogel W, Cox TM, Zoller H. 2011; Identification of mutations in SLC40A1 that affect ferroportin function and phenotype of human ferroportin iron overload. Gastroenterology. 140:2056–2063. 2063.e1. DOI: 10.1053/j.gastro.2011.02.064. PMID: 21396368.

5. Lelièvre P, Sancey L, Coll JL, Deniaud A, Busser B. 2020; Iron dysregulation in human cancer: altered metabolism, biomarkers for diagnosis, prognosis, monitoring and rationale for therapy. Cancers (Basel). 12:3524. DOI: 10.3390/cancers12123524. PMID: 33255972. PMCID: PMC7761132.

6. Xue D, Zhou CX, Shi YB, Lu H, He XZ. 2015; Decreased expression of ferroportin in prostate cancer. Oncol Lett. 10:913–916. Erratum in: Oncol Lett. 2021;21:257. DOI: 10.3892/ol.2015.3363. PMID: 26622594. PMCID: PMC4509077.

7. Deng Z, Manz DH, Torti SV, Torti FM. 2019; Effects of ferroportin-mediated iron depletion in cells representative of different histological subtypes of prostate cancer. Antioxid Redox Signal. 30:1043–1061. DOI: 10.1089/ars.2017.7023. PMID: 29061069. PMCID: PMC6354616.

8. Pinnix ZK, Miller LD, Wang W, D'Agostino R Jr, Kute T, Willingham MC, Hatcher H, Tesfay L, Sui G, Di X, Torti SV, Torti FM. 2010; Ferroportin and iron regulation in breast cancer progression and prognosis. Sci Transl Med. 2:43ra56. Erratum in: Sci Transl Med. 2010;2:46er1. DOI: 10.1126/scitranslmed.3001127. PMID: 20686179. PMCID: PMC3734848.

9. Deng L, Huang L, Guo Q, Shi X, Xu K. 2017; CREB1 and Smad3 mediate TGF-β3-induced Smad7 expression in rat hepatic stellate cells. Mol Med Rep. 16:8455–8462. DOI: 10.3892/mmr.2017.7654. PMID: 28983617.

10. Chen HC, Byrd JC, Muthusamy N. 2006; Differential role for cyclic AMP response element binding protein-1 in multiple stages of B cell development, differentiation, and survival. J Immunol. 176:2208–2218. DOI: 10.4049/jimmunol.176.4.2208. PMID: 16455977.

11. Barral S, Reitz C, Small SA, Mayeux R. 2014; Genetic variants in a 'cAMP element binding protein' (CREB)-dependent histone acetylation pathway influence memory performance in cognitively healthy elderly individuals. Neurobiol Aging. 35:2881.e7–2881.e10. DOI: 10.1016/j.neurobiolaging.2014.06.024. PMID: 25150575. PMCID: PMC4253058.

12. Wang P, Deng L, Zhuang C, Cheng C, Xu K. 2016; p-CREB-1 promotes hepatic fibrosis through the transactivation of transforming growth factor-β1 expression in rats. Int J Mol Med. 38:521–528. DOI: 10.3892/ijmm.2016.2630. PMID: 27279449.

13. Xin ZC, Hu HW, Lao ZH, Zhu LQ, Biskup E, Zhang HW. 2020; p-CREB-1 at Ser 133 is a potential marker for breast cancer. Eur Rev Med Pharmacol Sci. 24:11628–11638.
14. Xiao X, Zhang C, Grigoroiu-Serbanescu M, Wang L, Li L, Zhou D, Yuan TF, Wang C, Chang H, Wu Y, Li Y, Wu DD, Yao YG, Li M. 2018; The cAMP responsive element-binding (CREB)-1 gene increases risk of major psychiatric disorders. Mol Psychiatry. 23:1957–1967. DOI: 10.1038/mp.2017.243. PMID: 29158582.

15. Nabokina SM, Ramos MB, Valle JE, Said HM. 2015; Regulation of basal promoter activity of the human thiamine pyrophosphate transporter SLC44A4 in human intestinal epithelial cells. Am J Physiol Cell Physiol. 308:C750–C757. Erratum in: Am J Physiol Cell Physiol. 2017;313:C473. DOI: 10.1152/ajpcell.00381.2014. PMID: 25715703. PMCID: PMC4420793.
16. Teng X, Liu YY, Teng W, Brent GA. 2018; COUP-TF1 modulates thyroid hormone action in an embryonic stem-cell model of cortical pyramidal neuronal differentiation. Thyroid. 28:667–678. DOI: 10.1089/thy.2017.0256. PMID: 29205104. PMCID: PMC5952340.

17. Fischer SF, Schmidt K, Fiedler N, Glebe D, Schüttler C, Sun J, Gerlich WH, Repp R, Schaefer S. 2006; Genotype-dependent activation or repression of HBV enhancer II by transcription factor COUP-TF1. World J Gastroenterol. 12:6054–6058. DOI: 10.3748/wjg.v12.i37.6054. PMID: 17009409. PMCID: PMC4124418.

18. Yamaguchi H, Zhou C, Lin SC, Durand B, Tsai SY, Tsai MJ. 2004; The nuclear orphan receptor COUP-TFI is important for differentiation of oligodendrocytes. Dev Biol. 266:238–251. DOI: 10.1016/j.ydbio.2003.10.038. PMID: 14738874.

19. Rodriguez-Tirado C, Kale N, Carlini MJ, Shrivastava N, Rodrigues AA, Khalil BD, Bravo-Cordero JJ, Hong Y, Alexander M, Ji J, Behbod F, Sosa MS. 2022; NR2F1 is a barrier to dissemination of early-stage breast cancer cells. Cancer Res. 82:2313–2326. DOI: 10.1158/0008-5472.CAN-21-4145. PMID: 35471456. PMCID: PMC9203932.

20. Perets R, Kaplan T, Stein I, Hidas G, Tayeb S, Avraham E, Ben-Neriah Y, Simon I, Pikarsky E. 2012; Genome-wide analysis of androgen receptor targets reveals COUP-TF1 as a novel player in human prostate cancer. PLoS One. 7:e46467. DOI: 10.1371/journal.pone.0046467. PMID: 23056316. PMCID: PMC3464259.

21. Hunger SP, Ohyashiki K, Toyama K, Cleary ML. 1992; Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes Dev. 6:1608–1620. DOI: 10.1101/gad.6.9.1608. PMID: 1516826.

22. Wahlestedt M, Ladopoulos V, Hidalgo I, Sanchez Castillo M, Hannah R, Säwén P, Wan H, Dudenhöffer-Pfeifer M, Magnusson M, Norddahl GL, Göttgens B, Bryder D. 2017; Critical modulation of hematopoietic lineage fate by hepatic leukemia factor. Cell Rep. 21:2251–2263. DOI: 10.1016/j.celrep.2017.10.112. PMID: 29166614. PMCID: PMC5714592.

23. Waters KM, Sontag RL, Weber TJ. 2013; Hepatic leukemia factor promotes resistance to cell death: implications for therapeutics and chronotherapy. Toxicol Appl Pharmacol. 268:141–148. DOI: 10.1016/j.taap.2013.01.031. PMID: 23415677.

24. Xiang DM, Sun W, Zhou T, Zhang C, Cheng Z, Li SC, Jiang W, Wang R, Fu G, Cui X, Hou G, Jin GZ, Li H, Hou C, Liu H, Wang H, Ding J. 2019; Oncofetal HLF transactivates c-Jun to promote hepatocellular carcinoma development and sorafenib resistance. Gut. 68:1858–1871. DOI: 10.1136/gutjnl-2018-317440. PMID: 31118247.

25. Xiang DM, Sun W, Ning BF, Zhou TF, Li XF, Zhong W, Cheng Z, Xia MY, Wang X, Deng X, Wang W, Li HY, Cui XL, Li SC, Wu B, Xie WF, Wang HY, Ding J. 2018; The HLF/IL-6/STAT3 feedforward circuit drives hepatic stellate cell activation to promote liver fibrosis. Gut. 67:1704–1715. DOI: 10.1136/gutjnl-2016-313392. PMID: 28754776.

26. Inukai T, Hirose K, Inaba T, Kurosawa H, Hama A, Inada H, Chin M, Nagatoshi Y, Ohtsuka Y, Oda M, Goto H, Endo M, Morimoto A, Imaizumi M, Kawamura N, Miyajima Y, Ohtake M, Miyaji R, Saito M, Tawa A, et al. 2007; Hypercalcemia in childhood acute lymphoblastic leukemia: frequent implication of parathyroid hormone-related peptide and E2A-HLF from translocation 17;19. Leukemia. 21:288–296. DOI: 10.1038/sj.leu.2404496. PMID: 17183364.

27. Liu Q, Ge H, Liu P, Li Y. 2021; High Hepatic leukemia factor expression indicates a favorable survival in glioma patients. Medicine (Baltimore). 100:e23980. DOI: 10.1097/MD.0000000000023980. PMID: 33578515. PMCID: PMC7886392.

28. Kim BM, Song HS, Kim JY, Kwon EY, Ha SY, Kim M, Choi JH. 2022; Functional characterization of ABCA4 genetic variants related to Stargardt disease. Sci Rep. 12:22282. DOI: 10.1038/s41598-022-26912-6. PMID: 36566289. PMCID: PMC9790013.

29. Vivot A, Boutron I, Ravaud P, Porcher R. 2015; Guidance for pharmacogenomic biomarker testing in labels of FDA-approved drugs. Genet Med. 17:733–738. DOI: 10.1038/gim.2014.181. PMID: 25521333.

30. Cooper-DeHoff RM, Niemi M, Ramsey LB, Luzum JA, Tarkiainen EK, Straka RJ, Gong L, Tuteja S, Wilke RA, Wadelius M, Larson EA, Roden DM, Klein TE, Yee SW, Krauss RM, Turner RM, Palaniappan L, Gaedigk A, Giacomini KM, Caudle KE, et al. 2022; The clinical pharmacogenetics implementation consortium guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and statin-associated musculoskeletal symptoms. Clin Pharmacol Ther. 111:1007–1021. DOI: 10.1002/cpt.2557. PMID: 35152405. PMCID: PMC9035072.
31. Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, Griffiths AM, St George-Hyslop PH, Siminovitch KA. 2004; Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 36:471–475. DOI: 10.1038/ng1339. PMID: 15107849.

32. Leung E, Hong J, Fraser AG, Merriman TR, Vishnu P, Krissansen GW. 2006; Polymorphisms in the organic cation transporter genes SLC22A4 and SLC22A5 and Crohn's disease in a New Zealand Caucasian cohort. Immunol Cell Biol. 84:233–236. DOI: 10.1111/j.1440-1711.2006.01423.x. PMID: 16519742.

33. Martínez A, Martín MC, Mendoza JL, Taxonera C, Díaz-Rubio M, de la Concha EG, Urcelay E. 2006; Association of the organic cation transporter OCTN genes with Crohn's disease in the Spanish population. Eur J Hum Genet. 14:222–226. DOI: 10.1038/sj.ejhg.5201529. PMID: 16333318.

34. Tomer G, Wetzler G, Keddache M, Denson LA. 2009; Polymorphisms in the IBD5 locus are associated with Crohn disease in pediatric Ashkenazi Jewish patients. J Pediatr Gastroenterol Nutr. 48:531–537. DOI: 10.1097/MPG.0b013e318183138a. PMID: 19412005.

35. Jung ES, Park HJ, Kong KA, Choi JH, Cheon JH. 2017; Association study between OCTN1 functional haplotypes and Crohn's disease in a Korean population. Korean J Physiol Pharmacol. 21:11–17. DOI: 10.4196/kjpp.2017.21.1.11. PMID: 28066136. PMCID: PMC5214902.

36. Park HJ, Jung ES, Kong KA, Park EM, Cheon JH, Choi JH. 2016; Identification of OCTN2 variants and their association with phenotypes of Crohn's disease in a Korean population. Sci Rep. 6:22887. DOI: 10.1038/srep22887. PMID: 26965072. PMCID: PMC4786794.

37. Brookes MJ, Hughes S, Turner FE, Reynolds G, Sharma N, Ismail T, Berx G, McKie AT, Hotchin N, Anderson GJ, Iqbal T, Tselepis C. 2006; Modulation of iron transport proteins in human colorectal carcinogenesis. Gut. 55:1449–1460. DOI: 10.1136/gut.2006.094060. PMID: 16641131. PMCID: PMC1856421.

38. Liang B, Zhou C, Cui S, Lu H, Xu R, Xue D, Zou S, He X. 2021; Upregulation of miR-18a-5p promotes the proliferation of prostate cancer via inhibiting the expression of SLC40A1. Pathol Res Pract. 224:153448. DOI: 10.1016/j.prp.2021.153448. PMID: 34098197.

39. Wu J, Zhang L, Wu S, Yi X, Liu Z. 2020; miR-194-5p inhibits SLC40A1 expression to induce cisplatin resistance in ovarian cancer. Pathol Res Pract. 216:152979. DOI: 10.1016/j.prp.2020.152979. PMID: 32534701.

40. Gasparetto M, Pei S, Minhajuddin M, Stevens B, Smith CA, Seligman P. 2019; Low ferroportin expression in AML is correlated with good risk cytogenetics, improved outcomes and increased sensitivity to chemotherapy. Leuk Res. 80:1–10. DOI: 10.1016/j.leukres.2019.02.011. PMID: 30852438.

Fig. 1
The effect of
SLC40A1 promoter haplotypes or variants on its promoter activity. Luciferase activities were measured 48 h after transfection of reporter plasmids containing SLC40A1 haplotypes (A), or its variants (B) into HCT-116 cells. Relative luciferase activity of each haplotype or variant was compared to that of H1 or WT, respectively. Data shown represent mean ± SD from triplicate wells in a representative experiment. SLC40A1, solute carrier 40A1; WT, wild type. *p < 0.05, **p < 0.01, ***p < 0.001.
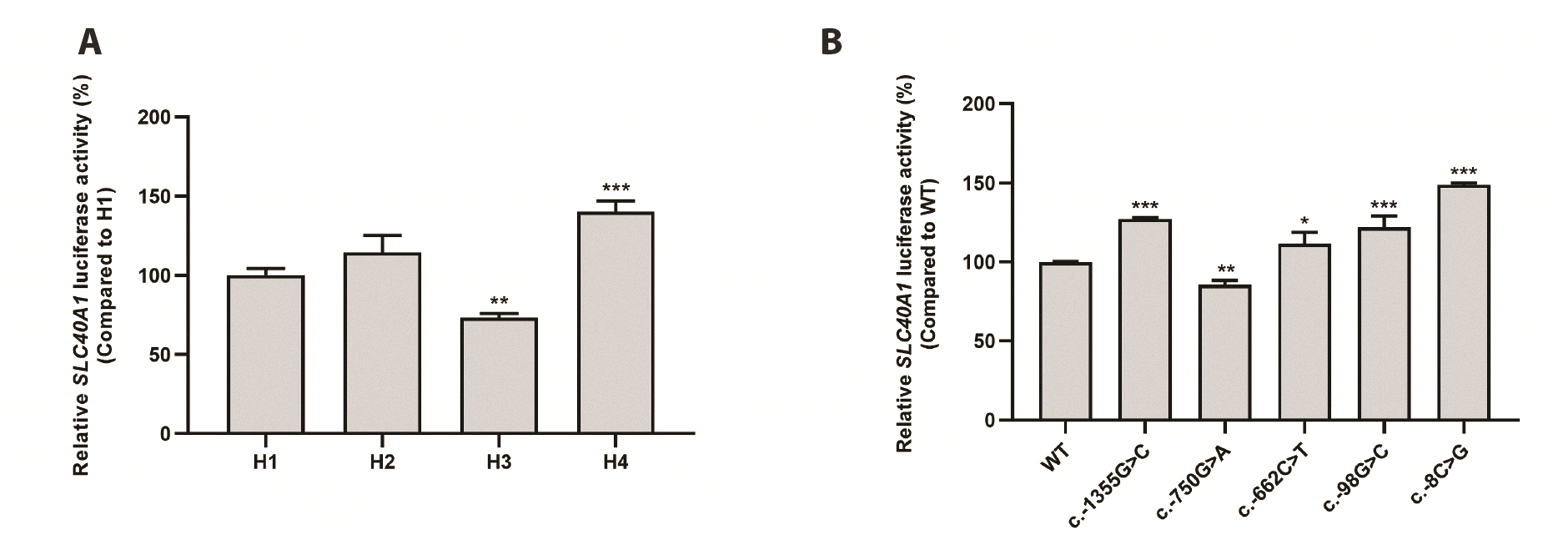
Fig. 2
DNA-protein binding assays to examine the interaction between
SLC40A1 promoter variants and transcription factors. Nuclear extracts from HCT-116 cells interacted with biotinylated oligonucleotide of CREB-1 consensus, c.-662C WT, or c.-662T variant (A) and biotinylated oligonucleotide COUP-TF1 consensus, c.-98G WT, or c.-98C variant (B) and biotinylated oligonucleotide of HLF consensus, c.-8C WT, or c.-8G variant (C). DNA-protein binding assay was performed using antibodies against CREB-1, COUP-TF1, or HLF. Data shown represent mean ± SD from three independent experiments. SLC40A1, solute carrier 40A1; WT, wild type; CREB-1, cAMP response element-binding protein-1; COUP-TF1, chicken ovalbumin upstream promoter transcription factor 1; HLF, hepatic leukemia factor; OD, optical density. *p < 0.05, **p < 0.01 vs. WT.
Fig. 3
The effect of transcription factors on the promoter activity of
SLC40A1. Luciferase activities were measured 48 h after co-transfection of SLC40A1 WT or its variants and various amounts of CREB-1 (A), COUP-TF1 (B), or HLF (C) cDNA into HCT-116 cells. The luciferase activity of each construct was compared with naïve promoter activity. Data shown represent mean ± SD from triplicate wells in a representative experiment. SLC40A1, solute carrier 40A1; WT, wild type; CREB-1, cAMP response element-binding protein-1; COUP-TF1, chicken ovalbumin upstream promoter transcription factor 1; HLF, hepatic leukemia factor. **p < 0.01, ***p < 0.001 vs. naïve promoter activity.
Table 1
Frequencies of SLC40A1 genetic variations in the promoter region
Table 2
Frequencies of SLC40A1 major haplotypes in the promoter region




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download