Abstract
The clustered regularly interspaced short palindromic repeats (CRISPR) system, a rapidly advancing genome editing technology, allows DNA alterations into the genome of organisms. Gene editing using the CRISPR system enables more precise and diverse editing, such as single nucleotide conversion, precise knock-in of target sequences or genes, chromosomal rearrangement, or gene disruption by simple cutting. Moreover, CRISPR systems comprising transcriptional activators/repressors can be used for epigenetic regulation without DNA damage. Stem cell DNA engineering based on gene editing tools has enormous potential to provide clues regarding the pathogenesis of diseases and to study the mechanisms and treatments of incurable diseases. Here, we review the latest trends in stem cell research using various CRISPR/Cas technologies and discuss their future prospects in treating various diseases.
Gene editing technology has evolved from the 1st generation zinc finger nucleases and 2nd generation transcription activator-like effector nucleases (TALEN) to the 3rd generation clustered regularly interspaced short palindromic repeats (CRISPR) system. Among them, CRISPR is a genome editing technology derived from bacterial adaptive immunity. Cas protein is programmed to cleave the target DNA following single guide RNA (sgRNA) (1). Target sequences induce double-strand breaks (DSBs) in the DNA and introduce insertion and deletion (indel) mutations that enable cell line or animal modeling via the non-homologous end joining (NHEJ) pathway (2, 3). The donor DNA is processed together, a specific sequence can be knocked in through homology-directed repair (HDR), another mechanism of DNA mismatch repair (4, 5). Furthermore, the expression level of the targeted gene can be regulated by binding a transcriptional repressor or activator to dead Cas (dCas), a Cas protein incapable of DNA cleavage (6, 7). Gaudelli et al. (8) and Komor et al. (9) proposed a base editor that could replace a specific base pair (C∙G to T∙A or A∙T to G∙C) without causing a DSB or externally adding a donor DNA. In 2019, this group also introduced a prime editor that can induce various mutations by inserting a desired sequence into the target position using reverse transcriptase (RT) (10). The development of such gene editing technology has made it possible to introduce and correct various mutations in the DNA sequence of organisms.
Stem cells are the earliest cell type in the cell lineage that can continuously proliferate and differentiate into various cell types (11). Embryonic stem cells (ESCs) with pluripotency are isolated from the inner cell mass of the blastocyst and are capable of self-renewal and differentiation into other specific cell lineages (11). Even in adults, the stem cells exist in different forms, such as intestinal stem cells (ISCs) that can differentiate into mature cell types necessary for normal intestinal functions and hematopoietic stem cells (HSCs) that can produce blood and immune-related cells (12, 13). However, since adult stem cells can differentiate into the cells of a specific lineage, their differentiation capacity is limited. To solve this problem, in 2006, Takahashi and Yamanaka (14) established a mechanism for differentiating mouse fibroblasts into induced pluripotent stem cells (iPSCs) by regulating the expression of Oct3/4, Sox2, c-Myc, and Klf4 genes. Since patient-specific iPSC production is achievable through this approach, disease modeling has become possible in recent years. This system is expected to be used in patient-specific drug screening or transplantation for cell therapy without triggering an immune response (15). This review describes CRISPR-based gene editing technology used in stem cells and its limitations and potential to be exploited for disease treatment (Fig. 1).
In 1987, CRISPR was first identified in Escherichia coli with the discovery of short tandem repeats interfering with the sequence (16). Since then, through rapid research, development and evolution over the past decade, CRISPR-based genome editing was finally demonstrated in human cells in 2013 (17-19). Here, we provide information on advanced genome editing technologies based on CRISPR (Fig. 2).
The CRISPR/Cas system was first identified in 2012 as an adaptive immune system that responds to virus invasion in bacteria by the research team of Jinek et al. (1). The CRISPR/Cas system consists of two components: a Cas nuclease and a sgRNA of 18∼20 bp that guides Cas to the desired DNA location. sgRNA consists of a tracrRNA binding sequence for Cas binding and crRNA complementary to the target sequence (20). SpCas9 derived from Streptococcus pyogenes, a representative Cas nuclease, is widely used for its convenience and high efficiency. In addition, Cas12a (Cpf1) recognizes crRNA-induced T-rich PAM and induces sticky-ended DSB, while Cas13 (C2C2) can edit single-stranded RNA targets (21-23).
The Cas9/sgRNA complex binds to the target sequence and the DNA-RNA hybridization by the sgRNA generates an R loop (24). After forming the R loop, the target and non-target strands are cleaved by the HNH and RuvC domain of SpCas9, respectively, causing DSB (25). DSB occurrence in the cell initiates various DNA repair processes. NHEJ mainly occurs in mammalian cells, which induces the insertion or deletion of random sequences to generate mutations causing frameshift with a probability of 2 out of 3 (2, 3). Cells also repair DSBs by copying and importing intact alleles via the HDR pathway for correct DNA repair (2, 3). HDR is induced at the target site by simultaneously processing the CRISPR system and donor DNA, having the desired nucleotide sequence enabling accurate knock-in. Alternatively, if a short micro-homologous sequence (5∼25 nucleotides) is there on each strand of the DSB-generating sequence, deletions or insertions of various sizes in the target sequence can be achieved through the microhomology-mediated end joining repair process (26). Through various DNA repair processes, this system can easily produce knock-out or knock-in cell lines in target genes. However, it is essential to solve the complications associated with off-target effects and immune response to use CRISPR/Cas system as a therapeutic agent. Of these, the ribonucleoprotein (RNP) complex of CRISPR/Cas is commonly used to reduce off-target effects. RNP, a complex of Cas protein and sgRNA, works rapidly in vivo for gene editing and reduces off-target effects due to its short half-life compared to other systems (27-29). Furthermore, efforts have been made to reduce off-target effects using truncated sgRNAs, engineered Cas variants, or allosterically regulated Cas systems (30). In addition, various anti-CRISPR (Acr) proteins have been identified from phages to inhibit the CRISPR system and are used to reduce the immune response (30-33). They block the normal role of Cas protein by interfering with Cas protein and crRNA binding, inhibiting DNA binding, or losing its ability of DNA cleavage (34-36).
As such, the CRISPR/Cas system can be applied to a wide range of research fields because it is easy to edit the target genome. In this review, we introduce various CRISPR/Cas systems and provide information on how they have been applied to stem cell research.
Although CRISPR/Cas has made many advances in gene editing, it can nevertheless result in unexpected indels, translocations, and chromosomal rearrangements caused by DSBs in non-dividing cells (37-41). A new alternative genome editing tool, the base editor system, was developed to overcome these limitations (42).
The base editor systems, including cytosine base editor (CBE) and adenine base editor (ABE), were introduced by Komor et al. (9) as a CRISPR system that was capable of allowing substitution at the single nucleotide level without inducing DSB. CBE comprises deaminase and nickase (nCas)9 (D10A) and can introduce nucleotide-level mutations from C∙G to T∙A in the target sequence (9, 43, 44). Cytosine deaminase replaces C with U in the base editing window, and nCas9, with the D10A mutation, nicks the non-target strand. Further, the complementary G is replaced with an A through cellular mismatch repair. Finally, during DNA replication or repair, U is replaced by T. Uracil glycosylase inhibitor was added to the CBE system to prevent the conversion of U to C by innate uracil DNA-glycosylase (9). CBE induces a C-to-T mutation in an editing window located approximately 4 to 8 bases from the distal end of the PAM in the 20 bp protospacer. CBE has reported to be used with various types of deaminases, such as APOBEC1, APOBEC3A, CDA1, and FERNY (42). These systems are used as base editing tools with improved functions to change the target window and increase efficiency.
The ABE system has the same base editing window as CBE and can introduce A∙T to G∙C mutations. The popularity of the ABE system has increased as it can treat nearly 50% of all human pathogenic point mutations (8, 45, 46). It comprises an engineered version of adenine deaminase (TadA) and nCas9 (D10A). In this system, adenine deaminase replaces A with I, which is changed to G during DNA repair or replication (8).
Various attempts have been made using base editing systems to broaden the primary editing window or narrow the editing window to the single nucleotide level within the target sequences while increasing editing efficiency (43). ABE8e with advanced TadAmax was shown to minimize the size of ABE with increased efficiency, while ABE9 (or NG-ABE9e: This system recognizes NG PAM sequences instead of NGG PAM sequences) changes one target A in editing windows (47-50). In addition, several studies have been conducted using cytosine and adenine deaminase in one base editor (SPACE, Target-ACEmax, and A&C-BEmax) (51-53). Recently, CGBE has been reported to have the capability of base substitution from C∙G to G∙C (54, 55). Also, using different PAM sequences of various Cas orthologous such as SpCas9 engineering version (NG and NGA), SaCas9 [NNGRR(T)], and Cpf1 (TTTV) was shown to extend the target sites (21, 56, 57).
About 50% of known human pathogenic mutation variants are point mutations (58). With the development of base editing technology, point mutations can be easily and efficiently introduced at the nucleotide level, which was difficult with the existing CRISPR gene editing method due to low knock-in efficiency.
Prime editor was reported by Anzalone et al. (10) in 2019, which has the capability to induce small insertion/deletion mutations and all nucleotide conversions. Prime editor comprises nCas9 carrying H840A variant and RT and works with pegRNA containing a spacer, primer-binding site (PBS), and RT template. When the spacer is combined with the target DNA sequence resulting in a nick on the non-target strand, PBS binds to the 3’ of the non-target strand, and RT synthesizes the complementary sequence along the RT template containing the desired mutant sequence. A 3’ edited DNA flap is created as the pegRNA is shed, which competes with the 5’ non-edited DNA flap. In addition, the prime editor uses nicking sgRNA to induce cleavage in the unedited DNA strand to increase DNA editing efficiency.
The prime editor is advantageous since it can introduce various mutations regardless of the type of editing since the RT template is copied and imported as it is (10). Recently, it was found that the efficiency of the prime editor could be increased by grafting chromatin-modulating peptide to facilitate access to target DNA sequences. Further, two prime editors can be used to induce large-scale deletions or chromosomal rearrangements (59, 60). Moreover, a new improved version of PEmax using Cas9 (R221K and N394K) variants, codon-optimized RT and mutation of the 34-aa linker with a bipartite SV40 NLS has been introduced. Additionally, the dominant negative MMR protein (MLH1dn) was transiently co-expressed with PEmax in cells to increase editing efficiency compared to the existing prime editor (61). Nelson et al. (62) improved prime editing efficiency using engineered pegRNA (epegRNA) with structured RNA motifs from tevopreQ1 or Mpknot for preventing degeneration and enhancing the stability of pegRNA. In conclusion, the development of the prime editor made it possible to apply more diverse genome editing beyond indel and limited base editing, which shows the scalability of the scope of use of gene editing.
Gilbert et al. (63) and Qi et al. (64) revealed that nuclease-null Cas9 (dCas9) does not cause DBS and binds to the promoter region in front of the transcription starting site, preventing RNA polymerase access and inhibits transcri-ption. Furthermore, a system was developed with the capability of more powerful transcriptional repression by including transcriptional repressors such as krüppel-associated box (KRAB), WRPW motif, or chromo shadow domain in the 3’ region of dCas9 (63, 65, 66). Thus, we can study gene functions and pathways with a knock-down strategy rather than permanent gene knock-out using the CRISPR interference (CRISPRi) system.
CRISPER activation (CRISPRa) increases target gene expression by conjugating various transcriptional activators to dCas9 that has lost the enzymatic activity. So far, dCas9-VP64 and various second-generation CRISPRa have been introduced to enhance gene overexpression activity (7, 63). In another report, the synergistic activation mediator (SAM) containing VP64 at the 5’ side of dCas and sgRNA with MS2 hairpin structure were used to induce the formation of four MCP-p65-HSF1 complexes at the target sequence, resulting in the strong transcriptional activity (67). Chavez et al. (68) introduced a method using a tripartite activator VP64-p65-Rta (VPR). Further, Tanenbaum et al. (69) developed Suntag, multiple protein-tagging systems using scFv with VP64.
Stem cells have self-renewal abilities, and specific types of cells like mesenchymal stromal cells have the capacity to release various factors, including VEGF, FGF, HGF, PGF, MCP-1, SDF-1, and Ang-1. In this respect, they can be used as promising materials in pathological mechanism, treatment, and regenerative medicine research by applying gene editing technology. Here, we summarize the studies that have employed gene-editing tools to stem cells for treating specific diseases (Table 1) (70-73).
ESCs are pluripotent stem cells with self-renewal capacity and the ability to differentiate into specific cell in the body. Here, we review several research cases exhibiting basic and treatment research by utilizing various gene editing technologies (74, 75) in ESCs.
The development of gene editing systems could greatly facilitate site-specific mutagenesis of human embryonic stem cells (hESCs), including introduction and modification of patient-specific mutations for disease modeling. Zhu et al. (76) demonstrated the strategy for knock-in GFP or RFP in a target gene without drug selection for both active and silent genes in hESCs using TALEN and CRISPRi. Zhou et al. (77) reported the generation of a lineage-specific hESC reporter cell line using the CRISPR/Cas9-system-mediated knock-in method.
Habib et al. (78) reported the development and application of inducible gene editing systems, such as iCas9, iCBE, iABE, and iPE2 to ESC. The possible therapeutic application was also demonstrated by repairing the E342K mutation in the SERPINA1 gene that causes α 1-antitrypsin (A1AT) deficiency in ESCs using the prime and base editors (78).
Kearns et al. (79) has constructed a lentivirus-based doxycycline-inducible CRISPRa/i system in ESCs (Table 2). Through this approach, dCas9-VP64 was used to induce the overexpression of SOX17, and dCas9-KRAB was used to suppress OCT4 to reveal the possibility of differentiation into other cell lineages (Table 2) (79).
Gene editing research in ESCs can be usefully applied to pathogenesis research and therapeutic development by elucidating the function of genes in the process of developing into each cell type and modeling genetic diseases.
Because iPSCs reprogram somatic cells from adult human, they can be used for patient-specific treatment or individual drug screening. In addition, iPSCs provide a way to achieve pluripotency without using embryos.
Park et al. (80) demonstrated mutation-correcting endothelial cell transplantation for the treatment of hemophilia for the first time. iPSCs derived from hemophilia patients with coagulation factor VIII gene mutations were restored to normalcy by CRISPR/Cas9 and subsequently differentiated into endothelial cells. Further, endothelial cells were transplanted into a hemophilia mouse model to examine its therapeutic effects.
Miki et al. (81) also demonstrated mucopolysaccharidosis-targeted gene editing in mouse iPSCs. Neomycin resistance was abolished by delivery of the CRIPSR/cas9 system along with donor DNA into iPSCs to remove a neomycin cassette in exon 6 of the defective α-L-iduronidase terminus.
Rett syndrome (RTT) is induced by mutations in methyl-CpG binding protein 2 (MECP2) and results in slow brain growth and intellectual disability (82). For RTT research, Le et al. (83) introduced the R270X mutation in the MECP2 gene in iPSCs with 20%∼30% efficiency by applying HDR based on the CRISPR system.
The Cys112Arg variant of apolipoprotein E4 (APOE4), a genetic risk factor for sporadic Alzheimer’s disease (sAD), was introduced into iPSCs using a CRISPR/Cas9-based knock-in method to investigate cell type-specific functions of APOE4 concerning AD pathology. The differentiated neuron cells with the APOE4 Cys112Arg variant, such as neurons, astrocytes, and microglia, resulted in extensive gene expression alterations and multiple cellular phenotypes potentially associated with AD pathogenesis (84).
Human urinary cells can be collected non-invasively and directly reprogrammed in iPSCs (85). Yang et al. (85) demonstrated CRISPR/Cas9-based HDR in urine-derived iPSCs from β-thalassemia patients with β-41/42 mutations to treat the mutation by inducing a TCTT deletion between the 41st and 42nd amino acids of HBB gene. This study is a meaningful result showing that the CRISPR system can be provided as a strategy for personalized treatment of β-thalassemia.
AGXT mutations cause excessive accumulation of oxalate in the liver and transport oxalate to the kidneys for-ming insoluble calcium oxalate, which results in primary hyperoxaluria type 1 (PH1) (86). Estève et al. (87) generated functionally corrected hepatocyte-like cells by knock-in the AGXT minigene fragment into AAVS1 locus from iPSCs of PH1 patients.
Mykkänen et al. (88), Vuorio et al. (89), Jalil et al. (90) used the base editing system for patient-derived iPSCs to rescue NOTCH3 (R133C) mutation, a dominant mutation in cerebral autosomal dominant arteriopathy with subcor-tical infarcts and leukoencephalopathy (CADASIL), and LDLR (R595Q) mutation, which causes familial hyper-cholesterolemia.
Chang et al. (91) succeeded in correcting the G2019S mutation in the LRRK2 gene, which is widely known as a mutation in Parkinson’s disease. They also found that ABE had higher on-target editing efficiency, lower off-targets, and indels than CRISPR/Cas9-based HDR in iPSCs.
Chemello et al. (92) used the CBE to replace T with C in the splicing donor sequence of Exon 50 in DMD to treat Duchenne muscular dystrophy (DMD) in iPSCs carrying the ΔExon 51 mutation in the DMD gene. Alternatively, two nucleotides were inserted into DMD Exon 52 of iPSCs using the prime editor. The edited cells were differentia-ted from cardiomyocyte, and the expression level of dystrophin was restored (92).
Spinal muscular atrophy (SMA) is an autosomal recessive motor neuromuscular disorder that weakens muscles and foils normal movement. Most patients with SMA are characterized by exon 7 skipping in the SMN2 transcript, producing unstable truncated proteins. Zhou et al. (93) inser-ted full-length SMN into SMA patient-derived iPSCs by inducing a targeted 9bp-deletion in the intronic splicing silencer-N1 of SMN2 with a prime editor. In the motor neurons derived from rescued iPSCs, apoptosis was reduced by restoring SMN protein. This result suggests that SMA disease caused by skipping exon 7 of the SMN2 gene can also be treated using the prime editor.
To induce neuronal differentiation, the expression of NEUROG2 and NEUROD1 was activated in iPSCs using dCas9-VP64 and dCas9-VPR (Table 2) (68). This study demonstrated the possibility of differentiating iPSCs to neurons through CRISPR-based gene expression activation, and in particular, showed that VPR system was more efficient than VP64 system.
Neural stem cells (NSCs) derived from iPSCs contain microRNA called the miR-199a/214 cluster, a negative regulator of hypoxia-induced cell migration. Luo et al. (94) suppressed the expression of micro RNA in iPSCs-derived NSCs using CRISPRi. This study has contributed to increasing the potential of NSCs for treating neurodegenerative diseases.
Adult stem cells have multipotency and can differentiate into several types of cells depending on their origin, such as HSCs, mesenchymal stem cells (MSCs), and NSCs (95). Because these stem cells are derived from adult tissue, they are suitable for targeting organ- or tissue-specific diseases.
Using the Cas nuclease, DNA cleavage can be easily induced at a desired location. The resulting random indel can change the amino acid sequence or induce a frameshift mutation to construct a knock-out model stem cell line. To understand the regulatory mechanism in mouse and human-derived hematopoietic stem and progenitor cells (HSPCs), functional studies have been reported by knocking out EED, SUZ12, and DNMT3A genes using Cas9/sgRNA RNPs (96, 97). AIDS, caused by HIV-1 infection, is generally treated using antiretroviral therapy (ART) (98). However, an effective treatment alternative is needed since ART requires long-term treatment, is expensive, and has various side effects. A few attempts have been made to induce resistance to infection by producing mutations in CCR5, a receptor through which HIV-1 enters. CD34+ HSPCs were treated and transplanted into humanized mice using the CRISPR system, and HIV-1 resistance was also confirmed in second transplantation (99). In addition, the lentiviral vector-mediated system was used to successfully increase resistance against HIV-1 infection in CD4+ T cells and HSPCs through the SaCas9, an easily deliverable small Cas ortholog (99, 100).
Long QT syndrome affects heart repolarization and increases QT length, resulting in abnormal heartbeats (101). Qi et al. (102) modeled an all-in-one episomal encoding base editors, such as epi-ABEmax, epi-ABEmax-NG, epi-AncBE4max, and epi-AncBE4max-NG in HSC to induce mutations in the representative LQT-related genes, KCNQ1 (L114P, R190Q), KCNH2 (Y616C, Y475C), or SCN5A (E1784K). In addition, using a base editor to generate stem cell lines harboring SCN5A (R1879W) mutant associated with Brugada syndrome showed that heart disease modeling is possible in stem cells (102, 103).
Diseases such as sickle cell disease (SCD) and β-tha-lassemia, caused by single nucleotide mutation, can be potential targets for stem cell gene editing. To treat SCD, Cas9 RNPs, and ssDNA donor nucleotides were used to edit a pathogenic point mutation located at the β-globin gene (HBB) of six different patient-derived CD34+ HSPCs (104). Also, Daniel Bauer’s group introduced BCL11A erythroid enhancer +58 C>T and HBB promoter −28 C>T mutations in SCD and β-thalassemia patient-derived HSCs using A3A (N57Q)-BE3 RNPs to ameliorate globin chain imbalance and red blood cell sickling with reduction of bystander mutation near the target nucleotide C (105). If HBB has an E6V (c.17A>T) mutation in SCD, ABE cannot reverse it to wild type. However, T∙A can be changed to C∙G, leading to non-pathogenic E6A, Hb-Ma-kassar (HBBG) (106). To apply these mutations, Chu et al. (107) tried using inlaid base editors to control and increase the base editing window efficiency. In addition, Chu et al. (107), Newby et al. (108), and Miller et al. (109) used a base editor-NRCH capable of non-G PAM (NRCH motif, H=A, C, T) targeting with increased targeting flexibility to edit human and mouse HSCs (A to G conversion, Val to Ala) and transplant them into mice to identify the therapeutic effect (108).
Mutations in SGCA (c.157G>A) cause limb-girdle muscular dystrophies, which weakens the shoulder and pelvic girdle muscles (110). In the study of Escobar et al. (110), the SGCA mutation (c.157G>A) that can induce exon ski-pping was corrected to normal by ABE treatment in muscle stem cells (MuSCs). This result showed that the expre-ssion of SGNA, which encodes α-sarcoglycan, was increa-sed in corrected MuSCs.
Since the use of MSCs avoids ethical issues and tumorigenesis, it is applicable for developing clinical applications including cell therapy and transplantation. The Furuhata et al. (111) induced differentiation from MSCs to white adipocyte-like cells using the target gene transcriptional activation system dCas9-SAM to induce programming of MSCs for therapeutic applications. This approach showed that overexpression of PPARG and CEBPA in MSCs could induce them to become beige adipocyte-like cells, suggesting that this system may also be applied to cell therapy (Table 2).
A few studies have shown gene editing in stem cell-derived organoids. Since stem cell-based organoids have the characteristics of each organ, they are advantageous for detecting diseases caused by genetic mutations. Schene et al. (112) used liver and intestinal organoids to mimic liver cancer growth by inducing a 6 bp deletion in CTNNB1. DGAT1 and ATP7B mutations in patients with congenital diarrhea and Wilson’s disease were restored in ISC organoids using prime editor (112).
Moreover, HDR, a more sophisticated DNA repair method in the CRISPR system, makes it possible to insert desired genes or mutations using donor DNA into precise locations. Schwank et al. (113) corrected ISC organoids derived from cystic fibrosis (CF) patients induced by the F508del mutation in CFTR to normal by a CRISPR system-mediated HDR technology. This group also performed cancer development modeling by applying the R175H, R249S, and Y220C mutations of TP53 using hepatocytes and colonic organoids via prime editing (114). This study demonstrated that the F580del and R785X mutations in CFTR that induce CF could be repaired using a prime editor in patient-derived gut organoids (114). Collectively, these research reports suggest that it is possible to efficiently introduce and correct mutations in 2D cells and 3D culturable organoids using various gene editing tools, which can be applied to various basic and therapeutic studies.
Stem cells are pluripotent and have significant advantages in cell differentiation and pathogenic studies. With the availability of single-cell analysis, lineage tracing has also become feasible. Recent advances in ex vivo and in vivo genome editing technologies have emerged as new therapeutic approaches with great potential for correcting genetic mutations in targeted stem cells. In this study, we summarized research trends about various CRISPR systems applied to stem cells. Not only gene knock-out/-in but also various types of editing, such as one nucleotide substitution and large insertion/deletion, are achievable through the CRISPR system. However, methods for precise editing as well as efficient and safe delivery without unwanted mutations in vitro and in vivo must still be developed before genome editing can be approved as a therapeutic tool. Moreover, there is still the off-target problem of editing sequences similar to the target, which needs to be addressed in the future for more sophisticated stem cell editing.
Recently, DNA DSBs introduced using Cas/sgRNA have been shown to cause deletions, inversions, and clone creations extending over many kilobases (115). Although difficulties in delivery and off-targets have been raised as limitations, CRISPR is still an irreplaceable and valuable tool for basic or clinical research. Furthermore, miniature Cas orthologs such as Cas12f have been discovered, enabling easier delivery (116). If various CRISPR systems are adequately utilized, it will significantly help stem cell research, such as lineage differentiation studies, disease modeling, and ex vivo treatment.
Notes
Funding
This work was supported by the Bio & Medical Tech-nology Development Program of the National Research Foundation (NRF) of Korea (NRF-2020M3A9D5A01082439 and NRF-2023R1A2C2004222).
Authors’ Contribution
Conceptualization: HL, KK. Data curation: DEY. Formal analysis: DEY, HL, KK. Funding acquisition: KK. Investi-gation: DEY, HL, KK. Methodology: DEY, HL, KK. Project administration: KK. Resources: KK. Software: KK. Supervision: HL, KK. Validation: HL, KK. Visualization: DEY. Writing – original draft: DEY. Writing – review and editing: HL, KK.
References
1. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012; A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337:816–821. DOI: 10.1126/science.1225829. PMID: 22745249. PMCID: PMC6286148.

2. Rouet P, Smih F, Jasin M. 1994; Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 14:8096–8106. DOI: 10.1128/MCB.14.12.8096. PMID: 7969147. PMCID: PMC359348.

3. Gallagher DN, Haber JE. 2018; Repair of a site-specific DNA cleavage: old-school lessons for Cas9-mediated gene editing. ACS Chem Biol. 13:397–405. DOI: 10.1021/acschembio.7b00760. PMID: 29083855. PMCID: PMC5835394.

4. van den Bosch M, Lohman PH, Pastink A. 2002; DNA double-strand break repair by homologous recombination. Biol Chem. 383:873–892. DOI: 10.1515/BC.2002.095. PMID: 12222678.
5. Song F, Stieger K. 2017; Optimizing the DNA donor template for homology-directed repair of double-strand breaks. Mol Ther Nucleic Acids. 7:53–60. DOI: 10.1016/j.omtn.2017.02.006. PMID: 28624224. PMCID: PMC5363683. PMID: e8da2e31beea42c686c5ffa612dadb80.

6. Marraffini LA, Sontheimer EJ. 2010; CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 11:181–190. DOI: 10.1038/nrg2749. PMID: 20125085. PMCID: PMC2928866.

7. Chavez A, Tuttle M, Pruitt BW, et al. 2016; Comparison of Cas9 activators in multiple species. Nat Methods. 13:563–567. DOI: 10.1038/nmeth.3871. PMID: 27214048. PMCID: PMC4927356.

8. Gaudelli NM, Komor AC, Rees HA, et al. 2017; Programmable base editing of A∙T to G∙C in genomic DNA without DNA cleavage. Nature. 551:464–471. Erratum in: Nature 2018;559:E8. DOI: 10.1038/nature24644. PMID: 29160308. PMCID: PMC5726555.

9. Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. 2016; Pro-grammable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 533:420–424. DOI: 10.1038/nature17946. PMID: 27096365. PMCID: PMC4873371.

10. Anzalone AV, Randolph PB, Davis JR, et al. 2019; Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 576:149–157. DOI: 10.1038/s41586-019-1711-4. PMID: 31634902. PMCID: PMC6907074.

11. Fortier LA. 2005; Stem cells: classifications, controversies, and clinical applications. Vet Surg. 34:415–423. DOI: 10.1111/j.1532-950X.2005.00063.x. PMID: 16266332.

12. Umar S. 2010; Intestinal stem cells. Curr Gastroenterol Rep. 12:340–348. DOI: 10.1007/s11894-010-0130-3. PMID: 20683682. PMCID: PMC2965634.

13. Dzierzak E, Bigas A. 2018; Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell. 22:639–651. DOI: 10.1016/j.stem.2018.04.015. PMID: 29727679.

14. Takahashi K, Yamanaka S. 2006; Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 126:663–676. DOI: 10.1016/j.cell.2006.07.024. PMID: 16904174.

15. Robinton DA, Daley GQ. 2012; The promise of induced pluripotent stem cells in research and therapy. Nature. 481:295–305. DOI: 10.1038/nature10761. PMID: 22258608. PMCID: PMC3652331.

16. Ishino Y, Krupovic M, Forterre P. 2018; History of CRISPR-Cas from encounter with a mysterious repeated sequence to genome editing technology. J Bacteriol. 200:e00580–17. DOI: 10.1128/JB.00580-17. PMID: 29358495. PMCID: PMC5847661.

17. Cho SW, Kim S, Kim JM, Kim JS. 2013; Targeted genome engi-neering in human cells with the Cas9 RNA-guided endo-nuclease. Nat Biotechnol. 31:230–232. DOI: 10.1038/nbt.2507. PMID: 23360966.

18. Cong L, Ran FA, Cox D, et al. 2013; Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. DOI: 10.1126/science.1231143. PMID: 23287718. PMCID: PMC3795411.

19. Mali P, Yang L, Esvelt KM, et al. 2013; RNA-guided human genome engineering via Cas9. Science. 339:823–826. DOI: 10.1126/science.1232033. PMID: 23287722. PMCID: PMC3712628.

20. Guo N, Liu JB, Li W, Ma YS, Fu D. 2022; The power and the promise of CRISPR/Cas9 genome editing for clinical application with gene therapy. J Adv Res. 40:135–152. DOI: 10.1016/j.jare.2021.11.018. PMID: 36100322. PMCID: PMC9481961.

21. Zetsche B, Gootenberg JS, Abudayyeh OO, et al. 2015; Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 163:759–771. DOI: 10.1016/j.cell.2015.09.038. PMID: 26422227. PMCID: PMC4638220.

22. Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. 2016; The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 532:517–521. DOI: 10.1038/nature17945. PMID: 27096362.

23. Abudayyeh OO, Gootenberg JS, Konermann S, et al. 2016; C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 353:aaf5573. DOI: 10.1126/science.aaf5573. PMID: 27256883. PMCID: PMC5127784.

24. Szczelkun MD, Tikhomirova MS, Sinkunas T, et al. 2014; Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A. 111:9798–9803. DOI: 10.1073/pnas.1402597111. PMID: 24912165. PMCID: PMC4103346.

25. Nishimasu H, Ran FA, Hsu PD, et al. 2014; Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 156:935–949. DOI: 10.1016/j.cell.2014.02.001. PMID: 24529477. PMCID: PMC4139937.

26. McVey M, Lee SE. 2008; MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 24:529–538. DOI: 10.1016/j.tig.2008.08.007. PMID: 18809224. PMCID: PMC5303623.

27. Farboud B, Jarvis E, Roth TL, et al. 2018; Enhanced genome editing with Cas9 ribonucleoprotein in diverse cells and orga-nisms. J Vis Exp. (135):57350. DOI: 10.3791/57350. PMID: 29889198. PMCID: PMC6101420.

28. Kim S, Kim D, Cho SW, Kim J, Kim JS. 2014; Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24:1012–1019. DOI: 10.1101/gr.171322.113. PMID: 24696461. PMCID: PMC4032847.

29. Martinez-Lage M, Puig-Serra P, Menendez P, Torres-Ruiz R, Rodriguez-Perales S. 2018; CRISPR/Cas9 for cancer therapy: hopes and challenges. Biomedicines. 6:105. DOI: 10.3390/biomedicines6040105. PMID: 30424477. PMCID: PMC6315587. PMID: 1072d8d705d44a0493545632fce87bd7.

30. Chapman JE, Gillum D, Kiani S. 2017; Approaches to reduce CRISPR off-target effects for safer genome editing. Appl Biosaf. 22:7–13. DOI: 10.1177/1535676017694148.

31. Shen CC, Hsu MN, Chang CW, et al. 2019; Synthetic switch to minimize CRISPR off-target effects by self-restricting Cas9 transcription and translation. Nucleic Acids Res. 47:e13. DOI: 10.1093/nar/gky1165. PMID: 30462300. PMCID: PMC6379646.

32. Marino ND, Pinilla-Redondo R, Csörgő B, Bondy-Denomy J. 2020; Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat Methods. 17:471–479. DOI: 10.1038/s41592-020-0771-6. PMID: 32203383. PMCID: PMC8510557.

33. Pawluk A, Davidson AR, Maxwell KL. 2018; Anti-CRISPR: discovery, mechanism and function. Nat Rev Microbiol. 16:12–17. DOI: 10.1038/nrmicro.2017.120. PMID: 29062071.

34. Zhu Y, Gao A, Zhan Q, et al. 2019; Diverse mechanisms of CRISPR-Cas9 inhibition by type IIC anti-CRISPR proteins. Mol Cell. 74:296–309.e7. DOI: 10.1016/j.molcel.2019.01.038. PMID: 30850331. PMCID: PMC6750902.

35. Harrington LB, Doxzen KW, Ma E, et al. 2017; A broad-spectrum inhibitor of CRISPR-Cas9. Cell. 170:1224–1233.e15. DOI: 10.1016/j.cell.2017.07.037. PMID: 28844692. PMCID: PMC5875921.

36. Bondy-Denomy J, Garcia B, Strum S, et al. 2015; Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature. 526:136–139. DOI: 10.1038/nature15254. PMID: 26416740. PMCID: PMC4935067.
37. Brunet E, Jasin M. 2018; Induction of chromosomal translocations with CRISPR-Cas9 and other nucleases: understanding the repair mechanisms that give rise to translocations. Adv Exp Med Biol. 1044:15–25. DOI: 10.1007/978-981-13-0593-1_2. PMID: 29956288. PMCID: PMC6333474.

38. Rose JC, Popp NA, Richardson CD, et al. 2020; Suppression of unwanted CRISPR-Cas9 editing by co-administration of catalytically inactivating truncated guide RNAs. Nat Commun. 11:2697. DOI: 10.1038/s41467-020-16542-9. PMID: 32483117. PMCID: PMC7264211. PMID: 32953ad84ae148f1a96180fcc11e3524.
39. Amendola M, Brusson M, Miccio A. 2022; CRISPRthripsis: the risk of CRISPR/Cas9-induced chromothripsis in gene therapy. Stem Cells Transl Med. 11:1003–1009. DOI: 10.1093/stcltm/szac064. PMID: 36048170. PMCID: PMC9585945.

40. Wen W, Zhang XB. 2022; CRISPR-Cas9 gene editing induced complex on-target outcomes in human cells. Exp Hematol. 110:13–19. DOI: 10.1016/j.exphem.2022.03.002. PMID: 35304271.

41. Lee J, Lim K, Kim A, et al. 2023; Prime editing with genuine Cas9 nickases minimizes unwanted indels. Nat Commun. 14:1786. DOI: 10.1038/s41467-023-37507-8. PMID: 36997524. PMCID: PMC10063541. PMID: d4e6a4c2bb8349c89d41ddb7631ab4af.

42. Thuronyi BW, Koblan LW, Levy JM, et al. 2019; Continuous evolution of base editors with expanded target compatibility and improved activity. Nat Biotechnol. 37:1070–1079. Erratum in: Nat Biotechnol 2019;37:1091. DOI: 10.1038/s41587-019-0193-0. PMCID: PMC6728210.

43. Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR. 2017; Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 35:371–376. DOI: 10.1038/nbt.3803. PMID: 28191901. PMCID: PMC5388574.
44. Komor AC, Zhao KT, Packer MS, et al. 2017; Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:a base editors with higher efficiency and product purity. Sci Adv. 3:eaao4774. DOI: 10.1126/sciadv.aao4774. PMID: 28875174. PMCID: PMC5576876.

45. Landrum MJ, Lee JM, Riley GR, et al. 2014; ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42:D980–D985. DOI: 10.1093/nar/gkt1113. PMID: 24234437. PMCID: PMC3965032.

46. Landrum MJ, Lee JM, Benson M, et al. 2016; ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44:D862–D868. DOI: 10.1093/nar/gkv1222. PMID: 26582918. PMCID: PMC4702865.

47. Richter MF, Zhao KT, Eton E, et al. 2020; Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol. 38:883–891. Erratum in: Nat Biotechnol 2020;38:901. DOI: 10.1038/s41587-020-0453-z. PMID: 32433547. PMCID: PMC7357821.

48. Rothgangl T, Dennis MK, Lin PJC, et al. 2021; In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels. Nat Biotechnol. 39:949–957. DOI: 10.1038/s41587-021-00933-4. PMID: 34012094. PMCID: PMC8352781.
49. Tu T, Song Z, Liu X, et al. 2022; A precise and efficient adenine base editor. Mol Ther. 30:2933–2941. DOI: 10.1016/j.ymthe.2022.07.010. PMID: 35821638. PMCID: PMC9481987.
50. Chen L, Zhang S, Xue N, et al. 2023; Engineering a precise adenine base editor with minimal bystander editing. Nat Chem Biol. 19:101–110. DOI: 10.1038/s41589-022-01163-8. PMID: 36229683.
51. Grünewald J, Zhou R, Lareau CA, et al. 2020; A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat Biotechnol. 38:861–864. DOI: 10.1038/s41587-020-0535-y. PMID: 32483364. PMCID: PMC7723518.
52. Sakata RC, Ishiguro S, Mori H, et al. 2020; Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat Biotechnol. 38:865–869. Erratum in: Nat Biotechnol 2020;38:901. DOI: 10.1038/s41587-020-0509-0.
53. Zhang X, Zhu B, Chen L, et al. 2020; Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat Biotechnol. 38:856–860. DOI: 10.1038/s41587-020-0527-y. PMID: 32483363.

54. Chen L, Park JE, Paa P, et al. 2021; Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat Commun. 12:1384. DOI: 10.1038/s41467-021-21559-9. PMID: 33654077. PMCID: PMC7925527. PMID: 179d414f933d4b028a782187beb2408e.
55. Sun N, Zhao D, Li S, Zhang Z, Bi C, Zhang X. 2022; Recon-structed glycosylase base editors GBE2.0 with enhanced C-to-G base editing efficiency and purity. Mol Ther. 30:2452–2463. DOI: 10.1016/j.ymthe.2022.03.023. PMID: 35381364. PMCID: PMC9263226.
56. Nishimasu H, Shi X, Ishiguro S, et al. 2018; Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 361:1259–1262. DOI: 10.1126/science.aas9129. PMID: 30166441. PMCID: PMC6368452.
57. Friedland AE, Baral R, Singhal P, et al. 2015; Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 16:257. DOI: 10.1186/s13059-015-0817-8. PMID: 26596280. PMCID: PMC4657203.
58. Rees HA, Liu DR. 2018; Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet. 19:770–788. Erratum in: Nat Rev Genet 2018;19:801. DOI: 10.1038/s41576-018-0059-1. PMID: 30323312. PMCID: PMC6535181.

59. Park SJ, Jeong TY, Shin SK, et al. 2021; Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor. Genome Biol. 22:170. DOI: 10.1186/s13059-021-02389-w. PMID: 34082781. PMCID: PMC8173820. PMID: 090e19d9eb3b4d668ab587bfea39465f.
60. Kweon J, Hwang HY, Ryu H, Jang AH, Kim D, Kim Y. 2023; Targeted genomic translocations and inversions generated using a paired prime editing strategy. Mol Ther. 31:249–259. DOI: 10.1016/j.ymthe.2022.09.008. PMID: 36114670. PMCID: PMC9840113.
61. Chen PJ, Hussmann JA, Yan J, et al. 2021; Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 184:5635–5652.e29. DOI: 10.1016/j.cell.2021.09.018. PMID: 34653350. PMCID: PMC8584034.
62. Nelson JW, Randolph PB, Shen SP, et al. 2022; Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol. 40:402–410. Erratum in: Nat Biotechnol 2022;40:432. DOI: 10.1038/s41587-021-01039-7. PMCID: PMC8930418.
63. Gilbert LA, Larson MH, Morsut L, et al. 2013; CRISPR-mediated modular RNA-guided regulation of transcription in euka-ryotes. Cell. 154:442–451. DOI: 10.1016/j.cell.2013.06.044. PMID: 23849981. PMCID: PMC3770145.
64. Qi LS, Larson MH, Gilbert LA, et al. 2013; Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 152:1173–1183. Erratum in: Cell 2021;184:844. DOI: 10.1016/j.cell.2013.02.022. PMID: 23452860. PMCID: PMC3664290.

65. Gilbert LA, Horlbeck MA, Adamson B, et al. 2014; Genome-scale CRISPR-mediated control of gene repression and activa-tion. Cell. 159:647–661. DOI: 10.1016/j.cell.2014.09.029. PMID: 25307932. PMCID: PMC4253859.
66. Konermann S, Brigham MD, Trevino A, et al. 2013; Optical control of mammalian endogenous transcription and epigenetic states. Nature. 500:472–476. DOI: 10.1038/nature12466. PMID: 23877069. PMCID: PMC3856241.

67. Konermann S, Brigham MD, Trevino AE, et al. 2015; Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 517:583–588. DOI: 10.1038/nature14136. PMID: 25494202. PMCID: PMC4420636.
68. Chavez A, Scheiman J, Vora S, et al. 2015; Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 12:326–328. DOI: 10.1038/nmeth.3312. PMID: 25730490. PMCID: PMC4393883.
69. Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. 2014; A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 159:635–646. DOI: 10.1016/j.cell.2014.09.039. PMID: 25307933. PMCID: PMC4252608.

70. Wei R, Yuan F, Wu Y, et al. 2018; Construction of a GLI3 compound heterozygous knockout human embryonic stem cell line WAe001-A-20 by CRISPR/Cas9 editing. Stem Cell Res. 32:139–144. DOI: 10.1016/j.scr.2018.09.010. PMID: 30278376.

71. Wang Z, Cui Y, Shan Y, et al. 2020; Generation of a MCPH1 knockout human embryonic stem cell line by CRISPR/Cas9 technology. Stem Cell Res. 49:102105. DOI: 10.1016/j.scr.2020.102105. PMID: 33370873.

72. Überbacher C, Obergasteiger J, Volta M, et al. 2019; Application of CRISPR/Cas9 editing and digital droplet PCR in human iPSCs to generate novel knock-in reporter lines to visualize dopaminergic neurons. Stem Cell Res. 41:101656. DOI: 10.1016/j.scr.2019.101656. PMID: 31733438. PMCID: PMC7322529.

73. Wu J, Hunt SD, Xue H, Liu Y, Darabi R. 2016; Generation and validation of PAX7 reporter lines from human iPS cells using CRISPR/Cas9 technology. Stem Cell Res. 16:220–228. DOI: 10.1016/j.scr.2016.01.003. PMID: 26826926. PMID: 7c6c805df9174335879ee8f80e3bdb95.
74. Lee J, Bayarsaikhan D, Bayarsaikhan G, Kim JS, Schwarz-bach E, Lee B. 2020; Recent advances in genome editing of stem cells for drug discovery and therapeutic application. Phar-macol Ther. 209:107501. DOI: 10.1016/j.pharmthera.2020.107501. PMID: 32061705.

75. Wu X, Jiang J, Gu Z, Zhang J, Chen Y, Liu X. 2020; Mesenchy-mal stromal cell therapies: immunomodulatory properties and clinical progress. Stem Cell Res Ther. 11:345. DOI: 10.1186/s13287-020-01855-9. PMID: 32771052. PMCID: PMC7414268. PMID: 500c255ba4ea4e5197bfeb6d27b44dde.
76. Zhu Z, Verma N, González F, Shi ZD, Huangfu D. 2015; A CRISPR/Cas-mediated selection-free knockin strategy in human embryonic stem cells. Stem Cell Reports. 4:1103–1111. DOI: 10.1016/j.stemcr.2015.04.016. PMID: 26028531. PMCID: PMC4471821. PMID: 7eefbed17c1f4feaa9f806e1d4c754bd.
77. Zhou J, Wang C, Zhang K, et al. 2016; Generation of human embryonic stem cell line expressing zsGreen in cholinergic neurons using CRISPR/Cas9 system. Neurochem Res. 41:2065–2074. DOI: 10.1007/s11064-016-1918-9. PMID: 27113041.

78. Habib O, Habib G, Hwang GH, Bae S. 2022; Comprehensive analysis of prime editing outcomes in human embryonic stem cells. Nucleic Acids Res. 50:1187–1197. DOI: 10.1093/nar/gkab1295. PMID: 35018468. PMCID: PMC8789035.

79. Kearns NA, Genga RM, Enuameh MS, Garber M, Wolfe SA, Maehr R. 2014; Cas9 effector-mediated regulation of transcri-ption and differentiation in human pluripotent stem cells. Development. 141:219–223. DOI: 10.1242/dev.103341. PMID: 24346702. PMCID: PMC3865759.
80. Park CY, Kim DH, Son JS, et al. 2015; Functional correction of large factor VIII gene chromosomal inversions in hemophilia A patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell. 17:213–220. DOI: 10.1016/j.stem.2015.07.001. PMID: 26212079.

81. Miki T, Vazquez L, Yanuaria L, et al. 2019; Induced pluripotent stem cell derivation and ex vivo gene correction using a mucopolysaccharidosis type 1 disease mouse model. Stem Cells Int. 2019:6978303. DOI: 10.1155/2019/6978303. PMID: 31065277. PMCID: PMC6466856. PMID: 64afa4737b334b2da07ac998db71302a.

82. Hagberg B, Aicardi J, Dias K, Ramos O. 1983; A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett's syndrome: report of 35 cases. Ann Neurol. 14:471–479. DOI: 10.1002/ana.410140412. PMID: 6638958.
83. Le TTH, Tran NT, Dao TML, et al. 2019; Efficient and precise CRISPR/Cas9-mediated MECP2 modifications in human- induced pluripotent stem cells. Front Genet. 10:625. DOI: 10.3389/fgene.2019.00625. PMID: 31333716. PMCID: PMC6614930. PMID: 24115b9b9c884f05a4f1cbce20d86fe4.
84. Lin YT, Seo J, Gao F, et al. 2018; APOE4 causes widespread molecular and cellular alterations associated with Alzheimer's disease phenotypes in human iPSC-derived brain cell types. Neuron. 98:1141–1154.e7. Erratum in: Neuron 2018;98: 1294. DOI: 10.1016/j.neuron.2018.05.008. PMID: 29861287. PMCID: PMC6023751.

85. Yang Y, Zhang X, Yi L, et al. 2016; Naïve induced pluripotent stem cells generated from β-thalassemia fibroblasts allow efficient gene correction with CRISPR/Cas9. Stem Cells Transl Med. 5:8–19. Erratum in: Stem Cells Transl Med 2016;5:267. DOI: 10.5966/sctm.2015-0157. PMID: 26676643. PMCID: PMC4704878.

86. Hoppe B, Beck BB, Milliner DS. 2009; The primary hyperoxalurias. Kidney Int. 75:1264–1271. DOI: 10.1038/ki.2009.32. PMID: 19225556. PMCID: PMC4577278.

87. Estève J, Blouin JM, Lalanne M, et al. 2019; Targeted gene therapy in human-induced pluripotent stem cells from a patient with primary hyperoxaluria type 1 using CRISPR/Cas9 technology. Biochem Biophys Res Commun. 517:677–683. DOI: 10.1016/j.bbrc.2019.07.109. PMID: 31402115.

88. Mykkänen K, Savontaus ML, Juvonen V, et al. 2004; Detection of the founder effect in Finnish CADASIL families. Eur J Hum Genet. 12:813–819. DOI: 10.1038/sj.ejhg.5201221. PMID: 15378071.

89. Vuorio AF, Aalto-Setälä K, Koivisto UM, et al. 2001; Familial hypercholesterolaemia in Finland: common, rare and mild mutations of the LDL receptor and their clinical consequences. Finnish FH-group. Ann Med. 33:410–421. DOI: 10.3109/07853890108995954. PMID: 11585102.

90. Jalil S, Keskinen T, Maldonado R, et al. 2021; Simultaneous high-efficiency base editing and reprogramming of patient fibroblasts. Stem Cell Reports. 16:3064–3075. DOI: 10.1016/j.stemcr.2021.10.017. PMID: 34822772. PMCID: PMC8693657.

91. Chang KH, Huang CY, Ou-Yang CH, et al. 2021; In vitro genome editing rescues parkinsonism phenotypes in induced pluripotent stem cells-derived dopaminergic neurons carrying LRRK2 p.G2019S mutation. Stem Cell Res Ther. 12:508. DOI: 10.1186/s13287-021-02585-2. PMID: 34551822. PMCID: PMC8456557. PMID: bc881b164425452fba2c024c95eb8485.

92. Chemello F, Chai AC, Li H, et al. 2021; Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci Adv. 7:eabg4910. DOI: 10.1126/sciadv.abg4910. PMID: 33931459. PMCID: PMC8087404.

93. Zhou M, Tang S, Duan N, et al. 2022; Targeted-deletion of a tiny sequence via prime editing to restore SMN expression. Int J Mol Sci. 23:7941. DOI: 10.3390/ijms23147941. PMID: 35887289. PMCID: PMC9317564. PMID: ec5edefa977b492892cca212b420c478.

94. Luo Y, Xu X, An X, Sun X, Wang S, Zhu D. 2016; Targeted inhibition of the miR-199a/214 cluster by CRISPR interference augments the tumor tropism of human induced pluripotent stem cell-derived neural stem cells under hypoxic condition. Stem Cells Int. 2016:3598542. DOI: 10.1155/2016/3598542. PMID: 27965712. PMCID: PMC5124688. PMID: 8422d3090a904c179f6c73c3d148551c.

95. Bozdağ SC, Yüksel MK, Demirer T. 2018; Adult stem cells and medicine. Adv Exp Med Biol. 1079:17–36. DOI: 10.1007/5584_2018_184. PMID: 29556955.
96. Brunetti L, Gundry MC, Kitano A, Nakada D, Goodell MA. 2018; Highly efficient gene disruption of murine and human hematopoietic progenitor cells by CRISPR/Cas9. J Vis Exp. (134):57278. DOI: 10.3791/57278. PMID: 29708546. PMCID: PMC5933422.

97. Gundry MC, Brunetti L, Lin A, et al. 2016; Highly efficient genome editing of murine and human hematopoietic progenitor cells by CRISPR/Cas9. Cell Rep. 17:1453–1461. DOI: 10.1016/j.celrep.2016.09.092. PMID: 27783956. PMCID: PMC5087995.

98. Moore RD, Chaisson RE. 1999; Natural history of HIV infection in the era of combination antiretroviral therapy. AIDS. 13:1933–1942. DOI: 10.1097/00002030-199910010-00017. PMID: 10513653.

99. Xu L, Yang H, Gao Y, et al. 2017; CRISPR/Cas9-mediated CCR5 ablation in human hematopoietic stem/progenitor cells confers HIV-1 resistance in vivo. Mol Ther. 25:1782–1789. DOI: 10.1016/j.ymthe.2017.04.027. PMID: 28527722. PMCID: PMC5542791.

100. Xiao Q, Chen S, Wang Q, et al. 2019; CCR5 editing by Staphylo-coccus aureus Cas9 in human primary CD4+ T cells and hematopoietic stem/progenitor cells promotes HIV-1 resistance and CD4+ T cell enrichment in humanized mice. Retrovirology. 16:15. Erratum in: Retrovirology 2019;16:20. DOI: 10.1186/s12977-019-0477-y. PMID: 31186067. PMCID: PMC6560749. PMID: daff0aaa003c4d039f406a0559f5988d.
101. Moss AJ. 2003; Long QT syndrome. JAMA. 289:2041–2044. DOI: 10.1001/jama.289.16.2041. PMID: 12709446.

102. Qi T, Wu F, Xie Y, et al. 2020; Base editing mediated generation of point mutations into human pluripotent stem cells for modeling disease. Front Cell Dev Biol. 8:590581. DOI: 10.3389/fcell.2020.590581. PMID: 33102492. PMCID: PMC7546412. PMID: fbb3a9afcbf246eda32aa4c8d22a846b.

103. Wilde AAM, Amin AS. 2018; Clinical spectrum of SCN5A mutations: long QT syndrome, Brugada syndrome, and cardio-myopathy. JACC Clin Electrophysiol. 4:569–579. DOI: 10.1016/j.jacep.2018.03.006. PMID: 29798782.
104. Dever DP, Bak RO, Reinisch A, et al. 2016; CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 539:384–389. DOI: 10.1038/nature20134. PMID: 27820943. PMCID: PMC5898607.

105. Zeng J, Wu Y, Ren C, et al. 2020; Therapeutic base editing of human hematopoietic stem cells. Nat Med. 26:535–541. DOI: 10.1038/s41591-020-0790-y. PMID: 32284612. PMCID: PMC7869435.

106. Sangkitporn S, Rerkamnuaychoke B, Sangkitporn S, Mitrakul C, Sutivigit Y. 2002; Hb G Makassar (beta 6:Glu-Ala) in a Thai family. J Med Assoc Thai. 85:577–582.
107. Chu SH, Packer M, Rees H, et al. 2021; Rationally designed base editors for precise editing of the sickle cell disease mutation. CRISPR J. 4:169–177. DOI: 10.1089/crispr.2020.0144. PMID: 33876959.

108. Newby GA, Yen JS, Woodard KJ, et al. 2021; Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature. 595:295–302. DOI: 10.1038/s41586-021-03609-w. PMID: 34079130. PMCID: PMC8266759.
109. Miller SM, Wang T, Randolph PB, et al. 2020; Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat Biotechnol. 38:471–481. DOI: 10.1038/s41587-020-0412-8. PMID: 32042170. PMCID: PMC7145744.

110. Escobar H, Krause A, Keiper S, et al. 2021; Base editing repairs an SGCA mutation in human primary muscle stem cells. JCI Insight. 6:e145994. DOI: 10.1172/jci.insight.145994. PMID: 33848270. PMCID: PMC8262330.

111. Furuhata Y, Nihongaki Y, Sato M, Yoshimoto K. 2017; Control of adipogenic differentiation in mesenchymal stem cells via endogenous gene activation using CRISPR-Cas9. ACS Synth Biol. 6:2191–2197. DOI: 10.1021/acssynbio.7b00246. PMID: 29077398.

112. Schene IF, Joore IP, Oka R, et al. 2020; Prime editing for functional repair in patient-derived disease models. Nat Com-mun. 11:5352. DOI: 10.1038/s41467-020-19136-7. PMID: 33097693. PMCID: PMC7584657. PMID: 0ba02ae845834a578eb870a005592a64.

113. Schwank G, Koo BK, Sasselli V, et al. 2013; Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 13:653–658. DOI: 10.1016/j.stem.2013.11.002. PMID: 24315439.

114. Geurts MH, de Poel E, Pleguezuelos-Manzano C, et al. 2021; Evalu-ating CRISPR-based prime editing for cancer modeling and CFTR repair in organoids. Life Sci Alliance. 4:e202000940. DOI: 10.26508/lsa.202000940. PMID: 34373320. PMCID: PMC8356249.

115. Kosicki M, Tomberg K, Bradley A. 2018; Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 36:765–771. DOI: 10.1038/nbt.4192. PMID: 30010673. PMCID: PMC6390938.

116. Harrington LB, Burstein D, Chen JS, et al. 2018; Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 362:839–842. DOI: 10.1126/science.aav4294. PMID: 30337455. PMCID: PMC6659742.

Fig. 1
Gene editing using clustered regularly interspaced short palindromic repeats (CRISPR) system in stem cells. CRISPR system could be used for various ex vivo or in vivo stem cell research, such as cell lineage or developmental study, generation of disease research, patient-specific pre-drug screening, and gene therapy. iPSCs: induced pluripotent stem cells, ESCs: embryonic stem cells, RNPs: ribonucleoproteins, AAV: adeno-associated virus.
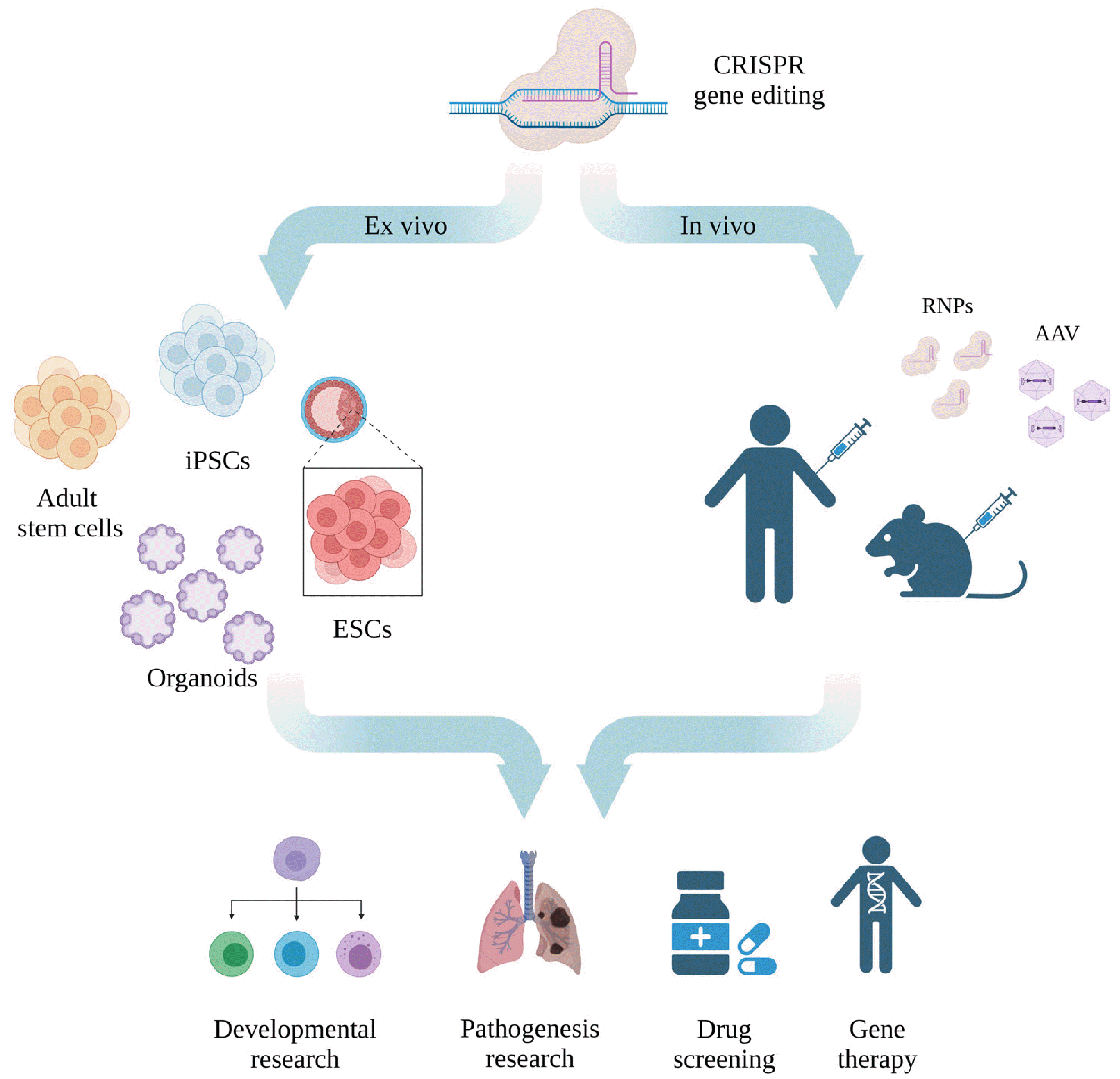
Fig. 2
Features and brief action mechanisms of diverse clustered regularly interspaced short palindromic repeats (CRISPR) systems. CRISPR system has been applied in various ways depending on the purpose. Cas nuclease makes random mutation at the cleavage site. The base editor can convert C to T or A to G. Prime editor could insert interested sequence through reverse transcriptase, and CRISPR activation/interference (CRISPRa/i) regulates specific gene expression. sgRNA: single guide RNA, ABE: adenine base editor, PBS: primer-binding site, RTT: Rett syndrome, RT: reverse transcriptase, nCas: nickase Cas, dCas: dead Cas, NHEJ: non-homologous end joining, HDR: homology-directed repair, CBE: cytosine base editor, UGI: uracil glycosylase inhibitor.
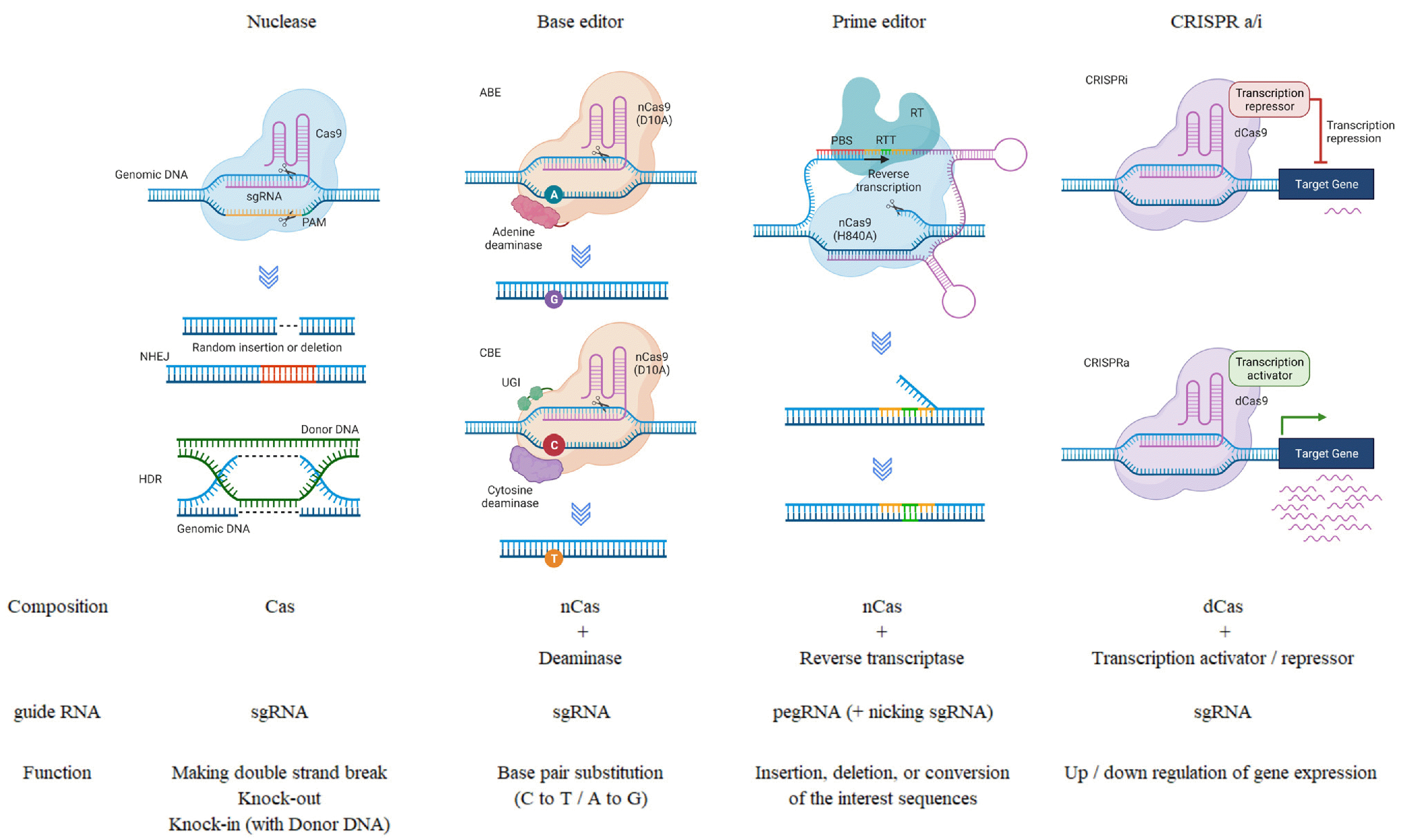
Table 1
Summary of diseases modeling and treatment studies applying various CRISPR/Cas systems to stem cells
| CRISPR type | Delivery method | Type of cells | Related disease | Target | Reference | ||
|---|---|---|---|---|---|---|---|
| Strategy | Form | Gene | Related mutation | ||||
| Knock-out | Plasmid | Electroporation | ESCs | Greig cephalopolysyndactylysyndrome, Pallister-Hall syndrome, preaxial polydactyly type IV, postaxial polydactyly type-A/B | GLI3 | Deletion in exon2 | (70) |
| Knock-out | Plasmid | Electroporation | ESCs | Primary autosomal recessive microcephaly and premature chromosome condensation syndrome | MCPH1 | 2 bp deletion in exon3 | (71) |
| Knock-in | Plasmid | Electroporation | ESCs | - | OCT4, PDX1 | OCT4-mOrange, OCT4-eGFP, PDX1-eGFP | (76) |
| Base editing, prime editing | Plasmid | Electroporation | ESCs | α-1 Antitrypsin deficiency | SERPINA1 | E342K | (78) |
| Knock-out | Plasmid | Lipid-based transfection | iPSCs | Hemophilia A | Coagulation factor VIII (F8) | int1/int22 inversion | (80) |
| Knock-in | Plasmid | Lipid-based transfection | iPSCs | Mucopolysaccharidosis type 1 | Idua | NeoR sequence insertion | (81) |
| Knock-in | Plasmid | Lipid-based transfection | iPSCs | Rett syndrome | MECP2 | R270X | (83) |
| Knock-in | Plasmid | Electroporation | iPSCs | Sporadic Alzheimer’s disease | APOE4 | R112C | (84) |
| Knock-in | Plasmid | Electroporation | iPSCs | β-Thalassemia | HBB | 4 bp deletion | (85) |
| Knock-in | mRNA | Lipid-based transfection | iPSCs | Primary hyperoxaluria type 1 | AAVS1 | AGXT minigene insertion | (87) |
| Knock-in | Plasmid | Electroporation | iPSCs | Parkinson’s disease | TH | eGFP insertion, | (72) |
| Knock-in | Plasmid | Electroporation | iPSCs | - | PAX7 | 2A-GFP insertion | (73) |
| Base editing | mRNA | Electroporation | iPSCs | Cerebral autosomaldominant arteriopathy with subcortical infarcts and leukoencephalopathy Familial hypercholesterolemia | NOTCH3 LDLR | R133C R595Q | (90) |
| Knock-in base editing | Plasmid | Electroporation | iPSCs | Parkinson’s disease | LRRK2 | G2019S | (91) |
| Base editing, prime editing | Plasmid | Electroporation | iPSCs | Duchenne muscular dystrophy | DMD | T>C (Ex50 SDS) 2 bp insertion (Ex52 reframing) | (92) |
| Prime editing | Plasmid | Lipid-based transfection | iPSCs | Spinal muscular atrophy | Intronic splicing silencer-N1 of SMN2 | 9 bp deletion | (93) |
| Knock-out | RNP | Electroporation | HSCs | - | DNMT3A | 58 bp deletion | (96) |
| Knock-out | Plasmid | Electroporation | HSCs | AIDS | CCR5 | Insertion or deletion | (98) |
| Base editing | Plasmid | Electroporation | HSCs | Long QT syndrome Brugada syndrome | KCNQ1 KCNH2 SCN5A | L114P/R190Q, Y616C/Y475C, E1784K/R1879W | (102) |
| Base editing | RNP | Electroporation | HSCs | β-Thalassemia Sickle cell disease | BCL11A erythroid enhancer HBB promoter | +58 C>T, −28 C>T | (105) |
| Base editing | mRNA | Electroporation | HSCs | Sickle cell disease | HBB | E6V | (107) |
| Base editing | mRNA /RNP | Electroporation | HSCs | Sickle cell disease | HBB | E6V | (108) |
| Base editing | Plasmid | Lipid-based transfection | iPSCs, MuSCs | Limb-girdle muscular dystrophies | SGCA | A53T | (110) |
| Prime editing | Plasmid | Electroporation | Liver and intestinal stem cell organoids | Liver cancer Congenital diarrhea Wilson disease | CTNNB1 DGAT1 ATP7B | 6 bp deletion, S210 deletion, S430fs | (112) |
| Knock-in | Plasmid | Lipid-based transfection | Intestinal stem cell organoids | Cystic fibrosis | CFTR | F508 deletion | (113) |
| Prime editing | Plasmid | Electroporation | Hepatocyte and colonic stem cell organoids Intestinal stem cell organoids | Hepatocellular carcinoma/ cancer Cystic fibrosis | TP53 CFTR | R175H/R249S/Y220C, F508del/R785X | (114) |
Table 2
Examples of gene regulation using CRISPRa/i system
| CRISPR system | Delivery method | Type of cells | Target | Reference | ||
|---|---|---|---|---|---|---|
| Strategy | CRISPR type | Gene | Differentiation | |||
| CRISPRa CRISPRi | dCas9-VP64 dCas9-KRAB | Lentivirus | ESCs | SOX17 OCT4 | - | (79) |
| CRISPRa | dCas9-SAM | Lentivirus | MSCs | PPARG, CEBPAP PARG, CEBPA, PRDM16 | White adipocyte-like cells Beige adipocyte-like cells | (111) |
| CRISPRa | dCas9-VP64 dCas9-VPR | Lentivirus | iPSCs | NEUROG2, NEUROD1 | Neuronal cells | (68) |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download