Abstract
This study aims to assess the quality and performance of predictive models for colorectal cancer liver metastasis (CRCLM). A systematic review was performed to identify relevant studies from various databases. Studies that described or validated predictive models for CRCLM were included. The methodological quality of the predictive models was assessed. Model performance was evaluated by the reported area under the receiver operating characteristic curve (AUC). Of the 117 articles screened, seven studies comprising 14 predictive models were included. The distribution of included predictive models was as follows: radiomics (n = 3), logistic regression (n = 3), Cox regression (n = 2), nomogram (n = 3), support vector machine (SVM, n = 2), random forest (n = 2), and convolutional neural network (CNN, n = 2). Age, sex, carcinoembryonic antigen, and tumor staging (T and N stage) were the most frequently used clinicopathological predictors for CRCLM. The mean AUCs ranged from 0.697 to 0.870, with 86% of the models demonstrating clear discriminative ability (AUC > 0.70). A hybrid approach combining clinical and radiomic features with SVM provided the best performance, achieving an AUC of 0.870. The overall risk of bias was identified as high in 71% of the included studies. This review highlights the potential of predictive modeling to accurately predict the occurrence of CRCLM. Integrating clinicopathological and radiomic features with machine learning algorithms demonstrates superior predictive capabilities.
Colorectal cancer (CRC) is a global health concern, ranking as the second most frequently diagnosed malignancy worldwide, with an incidence of 1.36 million cases each year [1,2]. Moreover, the global occurrence of CRC has been steadily rising, with an annual increase of 3.2% [3]. Metastases from CRC pose a significant obstacle to curative treatment, representing a pivotal factor contributing to CRC-related mortality [4]. Amongst the organs susceptible to CRC distant metastasis, the liver is the most frequently affected [5]. Population-based studies have revealed that 25% to 30% of CRC patients experience colorectal cancer liver metastases (CRCLM) throughout the course of the disease [6,7], as a result of lower gastrointestinal portal venous drainage [8]. While only 25% of patients with CRCLM qualify for operative resection [9], advancements in the field have expanded the treatment options for CRCLM. Early detection and accurate prediction of CRCLM are paramount to improving prognosis and delivering appropriate care for CRC patients.
The application of predictive modeling techniques in healthcare has brought about a transformative shift in the analysis and interpretation of medical data [10,11]. These advanced computational approaches offer the potential to uncover intricate patterns and relationships that may remain latent within large datasets, eluding traditional statistical methods [12]. Within the realm of CRCLM, the amalgamation of predictive modeling algorithms facilitates the development of robust prognostic tools that can consider the intricate interplay of multiple variables, providing individualized predictions [13-16]. These predictive models offer heightened accuracy by incorporating clinical parameters, pathological characteristics, and molecular biomarkers, empowering clinicians to make informed decisions on treatment strategies and patient management [17].
Despite numerous individual studies exploring the application of predictive models for CRCLM, a comprehensive evaluation of existing literature is currently lacking. A systematic analysis of the evidence is therefore timely.
This study was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA, 2020) guidelines [18]. A comprehensive literature search was conducted in May 2023 using the following terms: (“machine learning” OR “machine intelligen*” OR “machine vision*” OR “artificial intelligen*” OR “deep learning” OR “neural network” OR “supervised learning” OR “unsupervised learning” OR “reinforcement learning” OR “predictive model*” OR “predictive model*”) AND (“colon*” OR “rectal” OR “colorectal” OR “colonic” OR “rectum” OR “bowel” OR “intestine”) AND (“cancer*” OR “malignan*” OR “neoplas*” OR “tumor*” OR “tumour*”) AND (“liver meta*” OR “liver metastasis”) in the PubMed, MEDLINE, Embase, and Web of Science databases.
Studies were included if (1) the study population comprised male or female patients aged 18 years and above; (2) the participants consisted of adult individuals with CRC and liver metastasis; (3) the studies explicitly described and employed predictive modeling techniques to forecast the occurrence of CRCLM; (4) the studies reported the performance metrics of the predictive models, including sensitivity, specificity, accuracy, or the area under the receiver operating characteristic curve (AUC); (5) the full text of the articles was available for analysis; and (6) the articles were written in the English language. Case reports, reviews, and meta-analyses were excluded. The selection criteria did not specify a preferable study design or setting.
Two reviewers (YZ and BHA) screened the titles and abstracts of the articles identified in the search to identify potentially eligible studies. Full-text articles were obtained for all potentially eligible studies, and were independently reviewed by these two reviewers to determine eligibility. Any discrepancies were resolved with a third author (ISE) through discussion and consensus. The study selection process was documented using a PRISMA flow diagram.
The data collection was structured as per the Checklist for Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modeling Studies (CHARMS) [19]. The information extracted from each article included: the first author’s name, publication year, country of the study, study design, surgical approach, sample size, model details, model performance, and model evaluation. Due to significant heterogeneity observed in the study design, model development, and validation methodologies across the included studies, a meta-analysis was not performed.
The risk of bias (RoB) of the selected studies was assessed using the Prediction Model Risk of Bias Assessment Tool (PROBAST) [20]. This tool consists of four domains: patient selection, predictors, outcomes, and analysis. In addition, another evaluation was conducted to assess the applicability of the included studies across three domains: participants, predictors, and outcomes. Two independent reviewers (YZ and BHA) assessed the RoB for each domain, and assigned a rating of high, low, or unclear RoB. Any discrepancies were resolved through discussion with a third author (ISE). The RoB assessment was documented using a graphical summary.
Table 1 presents a summary of CRCLM predictive modeling techniques included in the review. In broad terms, predictive modeling encompasses statistical learning, machine learning, and deep learning approaches (Fig. 1) [21-23]. Statistical learning primarily involves developing and applying statistical methods and algorithms for predictive purposes. Notable examples of statistical learning methods include logistic regression (LR), least absolute shrinkage and selection operator (LASSO) regression, Cox regression, and nomogram. A nomogram represents a graphical tool employed within statistical learning to estimate the probability of an outcome or compute the value of a variable by considering the values of other associated variables [24]. Machine learning, a subfield of artificial intelligence (AI), is dedicated to creating algorithms and models that empower computers to learn from data and make predictions. Prominent machine learning techniques encompass support vector machine (SVM), decision tree, and random forest (RF). Deep learning, a subfield of machine learning, focuses on leveraging neural networks with multiple layers to comprehend intricate patterns and representations within data. The convolutional neural network (CNN) is a type of artificial neural network used in deep learning.
Radiomics is the extraction and analysis of numerous quantitative features from medical images, such as those obtained from computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography with computed tomography (PET-CT), into computationally exploitable information [25,26]. This method entails extracting textural, shape-related, intensity-based, and spatial attributes from images. By employing machine learning or statistical learning techniques on the derived radiomic features, prediction of clinical outcomes becomes possible.
The PRISMA flow diagram (Fig. 2) summarizes the literature screening process. After removing duplicates, the search strategy yielded 117 studies for full-text screening. Seven articles [15,27-32] were included in this systematic review. Table 2 summarizes the baseline characteristics of the included studies. All seven articles were retrospective in design, published between 2019 and 2022, and comprised six single-center studies and one multi-center study with a total of 35,989 participants. Notably, two studies had sample sizes larger than 2,000 patients. The distribution of included predictive models was as follows: radiomics (n = 3), LR (n = 3), Cox regression (n = 2), nomogram (n = 3), SVM (n = 2), RF (n = 2), and CNN (n = 2).
The predictive outcome of all included studies was the occurrence of CRCLM. Three studies integrated radiomics with LR, SVM, or RF; three employed nomograms in conjunction with LR or Cox regression; and two employed CNN (Table 2). The incorporation of clinicopathological variables as significant predictors was a consistent practice observed across all included studies. Amongst the clinicopathological variables used for predicting CRCLM, age (n = 5), sex (n = 3), carcinoembryonic antigen (CEA, n = 3), N stage (n = 4), and T stage (n = 3) emerged as the most frequently used predictors. Additional clinical predictors reported by individual studies included tumor size, tumor location, chemotherapy, CT images, digital pathological images, vascular emboli, lymphatic invasion, perineurial invasion, family history of CRCLM, and the presence of Kirsten rat sarcoma viral oncogene homologue (KRAS) mutations.
The discriminative performance of each predictive model was assessed using the AUC (Table 2). The AUC values, ranging from 0 to 1, were utilized to gauge the predictive performance of the models, with 0.5 denoting random chance, and 1.0 indicating a perfect fit [33]. AUC values surpassing 0.7 indicated a reasonably accurate prediction model [33]. Regarding the prediction of CRCLM, the mean AUCs ranged between 0.697 to 0.870, with 86% of models demonstrating clear discriminative ability (AUC > 0.70). Amongst the diverse models employed, the hybrid approach incorporating both clinical and radiomic features alongside SVM demonstrated the highest discriminative ability (AUC = 0.870). Other well-performing models included radiomics with SVM (AUC = 0.850), clinical features with RF (AUC = 0.860), the combined model integrating both clinical and radiomic features with RF (AUC = 0.860), as well as CNN model coupled with Cox regression and nomogram (AUC = 0.848).
All included studies conducted internal validation of their models through the resampling methods (Table 2), specifically cross-validation (n = 5) and/or bootstrapping (n = 4). Resampling techniques play a crucial role in evaluating and validating predictive models, ensuring their performance reliability [34]. Cross-validation partitions the available data into distinct subsets, or folds, where the model is trained on one fold (training set), and evaluated on the remaining fold (validation set) [35]. This technique mitigates overfitting risks and gauges the model’s generalizability across diverse data subsets. Conversely, bootstrapping involves generating multiple bootstrap samples through random sampling with replacement from the original dataset [35]. These bootstrap samples, of the same size as the original dataset, enable the training and evaluation of multiple models. Bootstrapping’s diverse resampled datasets facilitate the estimation of model performance variability and calculation of confidence intervals for key metrics, such as accuracy or AUC. The calibration, which pertains to the correspondence between the anticipated probabilities derived from a predictive model and the actual observed outcomes, signifying the precision and dependability of the model’s predictions, was reported for two nomogram models. None of the included studies reported external validation.
Fig. 3 and Supplementary Table 1, 2 present the results of RoB and applicability assessment. The overall RoB was determined to be high in 71% of the included studies. Within the participants domain, 57% of the studies were assessed as having a low RoB. However, one study [28] was identified as having a high RoB due to insufficiently detailed inclusion criteria, while two studies [30,31] were classified as having an unclear RoB for similar reasons. Regarding the domain of predictors, 43% of the studies were assessed as low RoB. Notably, four studies [15,27,29,30] that utilized radiomic features were assigned a high RoB, due to the inherent complexity associated with these features. As for the outcomes domain, 71% of the studies were assessed as low RoB, with two studies [15,29] identified as having a high RoB due to their relatively small sample sizes (< 100). For the domain of analysis, 71% of the studies were assessed as low RoB, while only one study [29] was deemed to have a high RoB due to a lack of information regarding the 95% confidence interval of AUC, and the absence of details concerning the predictor selection process, coupled with a small sample size.
This systematic review encompassed a comprehensive analysis of seven studies, collectively reporting 14 predictive models incorporating diverse risk factors for CRCLM. Our study sought to identify and evaluate the predictive models that exhibited promising discrimination capabilities for CRCLM. By considering the characteristics of the included studies, essential insights regarding the current research landscape of predictive modeling for CRCLM were obtained.
The retrospective design of all seven studies underscored the reliance on historical data to develop and validate predictive models. This approach allowed clinicians and researchers to leverage existing patient information to construct and assess the performance of the models [36]. Furthermore, all studies were published between 2019 and 2022, reflecting the recent interest in predictive modeling for CRCLM. The temporal proximity of these articles suggests an evolving and dynamic research landscape characterized by the ongoing pursuit of innovative strategies for predicting CRCLM.
The distribution of predictive models also exhibited considerable diversity with various statistical/machine/deep learning approaches, including LR, Cox regression, nomogram, SVM, RF, and CNN. The 14 predictive models demonstrated mean AUC values ranging from 0.697 to 0.870, with the majority (71%) achieving an AUC exceeding 0.75, indicating valuable discriminatory performance [37]. Three studies used radiomic features in conjunction with machine learning algorithms to predict CRCLM, with one study resulting in the highest AUC of 0.870 by combining clinical and radiomic features with SVM. This highlights the effectiveness of integrating multiple data sources and advanced computational techniques at enhancing the accuracy of CRCLM prediction.
CT, MRI, and PET-CT are frequently used imaging modalities to detect CRCLM. Nevertheless, their diagnostic sensitivity and accuracy can vary depending on equipment and reporting radiologist’s expertise [38]. In a meta-analysis with a 20-year study period, the detection sensitivities for CRCLM for CT, MRI, and PET-CT were reported as 74.4%, 80.3%, and 81.4%, respectively [39]. Radiomics has shown promise in surpassing the limitations of conventional imaging, by enabling quantitative and comprehensive analysis of tumor characteristics [40]. Its ability to capture hidden patterns and heterogeneity within the tumor microenvironment provides valuable insights into the risk and potential for CRCLM development. Our results showed that radiomic models employing quantitative image features extracted from CT or MRI achieved an AUC greater than 0.70. These findings are consistent with studies that used radiomic models to predict distant metastases in other types of cancer [41-43], suggesting that the combination of radiomic features and machine learning techniques has the potential to improve the predictive accuracy of CRCLM.
However, the application of radiomics in predicting CRCLM faces several challenges. One significant drawback is the potential variability in image features across healthcare settings. Variations in imaging protocols, equipment, and image acquisition techniques can lead to inconsistencies in the extracted radiomic features. As a result, a model developed and validated in one healthcare setting may not perform optimally when applied to another setting. To overcome the lack of generalizability, the integration of clinicopathological alongside radiomic characteristics becomes crucial.
Clinical attributes provide important contextual information that complements the image-based features captured by radiomics. While radiomic features may be influenced by site-specific variations, clinical attributes tend to have a more consistent definition. Age, sex, CEA levels, and tumor stage (T and N staging) were identified as the most widely used predictors for CRCLM. Age is a fundamental demographic feature that may serve as a proxy for multiple factors associated with disease pathogenesis and progression. Age-related alterations in the immune response and decreased immune surveillance, including impaired T-cell proliferation, increased CD8+ cytotoxic cell numbers, and decreased CD4+ T-cell and CD19+ B-cell numbers, have been postulated to affect the immune capacity to identify and eliminate metastatic CRC cells, thereby potentially contributing to an increased risk of CRCLM [44-49].
Similarly, sex disparities in metastatic CRC outcomes have been observed [50,51], establishing sex as a noteworthy predictor for CRCLM. Hormones, including estrogen and testosterone, have been implicated in CRC development and progression [52,53]. Sex-specific hormonal differences influence the susceptibility to CRCLM, as estrogen potentially exerts protective effects against CRCLM development [52,54]. CEA is associated with other key factors, such as large tumor size, advanced tumor stage, lymph node involvement, and its involvement in facilitating tumor cell adhesion, migration, and invasion processes [55-59]. The regular monitoring of CEA levels plays a pivotal role in identifying individuals at heightened risk of CRCLM, and aids in the formulation of appropriate surveillance and treatment strategies [60]. The depth of tumor invasion and lymph node involvement are also high-risk features and predictors for CRCLM development [61].
SVM, RF, and Cox regression with nomogram showed better performance than LR and CNN in the prediction of CRCLM. SVM can effectively handle complex and high-dimensional clinical and radiomic features, capturing intricate patterns and non-linear relationships, which is crucial for accurate predictions [62]. The ability of SVM to separate data points into different classes by finding an optimal hyperplane maximally distant from the data points of different classes enhances its predictive accuracy [63]. RF is an ensemble learning method that combines multiple decision trees to make predictions. By constructing a multitude of decision trees on random subsets of the data and aggregating their predictions, RF can mitigate overfitting, and improve the generalizability of the predictive model [64]. Cox regression with nomogram is a survival analysis technique that considers time-to-event data and covariates. By incorporating clinical variables and constructing a nomogram, which visually represents the contribution of each predictor, this approach allows estimation of individualized probabilities for developing CRCLM.
On the other hand, LR and CNN may exhibit comparatively lower prediction performance. LR assumes linear relationships between predictors and the outcome, which may not adequately capture the complex interactions and non-linear associations present. CNN, although powerful in image analysis tasks, may not fully exploit the relevant features for CRCLM prediction, as it primarily focuses on extracting spatial patterns from medical images instead of incorporating clinical variables. Incorporating CNN for digital pathological image analysis, followed by Cox regression and nomogram, can enhance the predictive accuracy of the model.
Despite the generally high accuracies observed in the predictive models assessed in this review, there remain several limitations. One notable shortcoming is the absence of external validation in all seven included studies. In addition, calibration, which provides information about the agreement between predicted probabilities and observed outcomes, was only reported in three studies (43%) that utilized nomograms. Poor calibration suggests potential under- or overestimation of the desired outcome by the model. Furthermore, the assessment of model performance primarily relied on discrimination measures, specifically AUC, as calibration measures were absent in four (57%) of the included studies. Due to the considerable heterogeneity amongst the included studies, conducting a pooled analysis or comparative meta-analysis of predictive models was not feasible.
Nonetheless, this review provides valuable insights into the current landscape of predictive models for CRCLM. Our findings highlight the potential of various algorithms by augmented by clinical and radiomic features in accurately predicting CRCLM. Future research could address the identified limitations by incorporating external validation in predictive model development. Efforts should also be made to improve the reporting of calibration measures, enhancing model performance and calibration accuracy. Furthermore, the heterogeneity of the included studies highlights the need for standardized methodologies and reporting guidelines in the field of predictive modeling. Developing consensus criteria and guidelines would facilitate more rigorous and comparable evaluations of predictive models, facilitating more robust evidence synthesis and meta-analyses. Collaborative efforts, particularly multi-center studies, are essential to enhance the generalizability and clinical utility of predictive models for CRCLM.
This review demonstrates the potential of predictive modeling for CRCLM. The integration of clinicopathological and radiomic features with machine learning algorithms showed superior predictive capabilities. External validation studies are necessary to establish the reliability and generalizability of predictive models, particularly across diverse healthcare settings. Improved reporting and standardized methodologies are also required to facilitate the integration of predictive models into routine clinical practice.
Supplementary data related to this article can be found at https://doi.org/10.14701/ahbps.23-078.
REFERENCES
1. Edwards BK, Ward E, Kohler BA, Eheman C, Zauber AG, Anderson RN, et al. 2010; Annual report to the nation on the status of cancer, 1975-2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 116:544–573. DOI: 10.1002/cncr.24760. PMID: 19998273. PMCID: PMC3619726.

2. Morgan E, Arnold M, Gini A, Lorenzoni V, Cabasag C, Laversanne M, et al. 2023; Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut. 72:338–344. DOI: 10.1136/gutjnl-2022-327736. PMID: 36604116.

3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. 2018; Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68:394–424. DOI: 10.3322/caac.21492. PMID: 30207593.

4. Yu X, Zhu L, Liu J, Xie M, Chen J, Li J. 2020; Emerging role of immunotherapy for colorectal cancer with liver metastasis. Onco Targets Ther. 13:11645–11658. DOI: 10.2147/OTT.S271955. PMID: 33223838. PMCID: PMC7671511.
5. Tauriello DV, Calon A, Lonardo E, Batlle E. 2017; Determinants of metastatic competency in colorectal cancer. Mol Oncol. 11:97–119. DOI: 10.1002/1878-0261.12018. PMID: 28085225. PMCID: PMC5423222.

6. Engstrand J, Nilsson H, Strömberg C, Jonas E, Freedman J. 2018; Colorectal cancer liver metastases-a population-based study on incidence, management and survival. BMC Cancer. 18:78. DOI: 10.1186/s12885-017-3925-x. PMID: 29334918. PMCID: PMC5769309.
7. Martin J, Petrillo A, Smyth EC, Shaida N, Khwaja S, Cheow H, et al. 2020; Colorectal liver metastases: current management and future perspectives. World J Clin Oncol. 11:761. DOI: 10.5306/wjco.v11.i10.761. PMID: 33200074. PMCID: PMC7643190.

8. Hugen N, van de Velde CJH, de Wilt JHW, Nagtegaal ID. 2014; Metastatic pattern in colorectal cancer is strongly influenced by histological subtype. Ann Oncol. 25:651–657. DOI: 10.1093/annonc/mdt591. PMID: 24504447. PMCID: PMC4433523.

9. Ivey GD, Johnston FM, Azad NS, Christenson ES, Lafaro KJ, Shubert CR. 2022; Current surgical management strategies for colorectal cancer liver metastases. Cancers (Basel). 14:1063. DOI: 10.3390/cancers14041063. PMID: 35205811. PMCID: PMC8870224.

10. Yu KH, Beam AL, Kohane IS. 2018; Artificial intelligence in healthcare. Nat Biomed Eng. 2:719–731. DOI: 10.1038/s41551-018-0305-z. PMID: 31015651.

11. Jiang F, Jiang Y, Zhi H, Dong Y, Li H, Ma S, et al. 2017; Artificial intelligence in healthcare: past, present and future. Stroke Vasc Neurol. 2:230–243. DOI: 10.1136/svn-2017-000101. PMID: 29507784. PMCID: PMC5829945.

12. Secinaro S, Calandra D, Secinaro A, Muthurangu V, Biancone P. 2021; The role of artificial intelligence in healthcare: a structured literature review. BMC Med Inform Decis Mak. 21:125. DOI: 10.1186/s12911-021-01488-9. PMID: 33836752. PMCID: PMC8035061.

13. Vorontsov E, Cerny M, Régnier P, Di Jorio L, Pal CJ, Lapointe R, et al. 2019; Deep learning for automated segmentation of liver lesions at CT in patients with colorectal cancer liver metastases. Radiol Artif Intell. 1:180014. DOI: 10.1148/ryai.2019180014. PMID: 33937787. PMCID: PMC8017429.
14. Paredes AZ, Hyer JM, Tsilimigras DI, Moro A, Bagante F, Guglielmi A, et al. 2020; A novel machine-learning approach to predict recurrence after resection of colorecta liver metastases. Ann Surg Oncol. 27:5139–5147. DOI: 10.1245/s10434-020-08991-9. PMID: 32779049.

15. Taghavi M, Trebeschi S, Simões R, Meek DB, Beckers RC, Lambregts DM, et al. 2021; Machine learning-based analysis of CT radiomics model for prediction of colorectal metachronous liver metastases. Abdom Radiol (NY). 46:249–256. DOI: 10.1007/s00261-020-02624-1. PMID: 32583138.

16. Rompianesi G, Pegoraro F, Ceresa CD, Montalti R, Troisi RI. 2022; Artificial intelligence in the diagnosis and management of colorectal cancer liver metastases. World J Gastroenterol. 28:108–122. DOI: 10.3748/wjg.v28.i1.108. PMID: 35125822. PMCID: PMC8793013.

17. Visvikis D, Cheze Le Rest C, Jaouen V, Hatt M. 2019; Artificial intelligence, machine (deep) learning and radio (geno) mics: definitions and nuclear medicine imaging applications. Eur J Nucl Med Mol Imaging. 46:2630–2637. DOI: 10.1007/s00259-019-04373-w. PMID: 31280350.

18. Page MJ, Mckenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. 2021; The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Int J Surg. 88:105906. DOI: 10.1016/j.ijsu.2021.105906. PMID: 33789826.
19. Moons KG, de Groot JA, Bouwmeester W, Vergouwe Y, Mallett S, Altman DG, et al. 2014; Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med. 11:e1001744. DOI: 10.1371/journal.pmed.1001744. PMID: 25314315. PMCID: PMC4196729.

20. Moons KGM, Wolff RF, Riley RD, Whiting PF, Westwood M, Collins GS, et al. 2019; PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med. 170:W1–W33. DOI: 10.7326/M18-1377. PMID: 30596876.

21. Sidey-Gibbons JAM, Sidey-Gibbons CJ. 2019; Machine learning in medicine: a practical introduction. BMC Med Res Methodol. 19:64. DOI: 10.1186/s12874-019-0681-4. PMID: 30890124. PMCID: PMC6425557.

22. Pfob A, Lu SC, Sidey-Gibbons C. 2022; Machine learning in medicine: a practical introduction to techniques for data pre-processing, hyperparameter tuning, and model comparison. BMC Med Res Methodol. 22:282. DOI: 10.1186/s12874-022-01758-8. PMID: 36319956. PMCID: PMC9624048.

23. Bektaş M, Tuynman JB, Costa Pereira J, Burchell GL, Van Der Peet DL. 2022; Machine learning algorithms for predicting surgical outcomes after colorectal surgery: a systematic review. World J Surg. 46:3100–3110. DOI: 10.1007/s00268-022-06728-1. PMID: 36109367. PMCID: PMC9636121.

24. Jalali A, Alvarez-Iglesias A, Roshan D, Newell J. 2019; Visualising statistical models using dynamic nomograms. PloS One. 14:e0225253. DOI: 10.1371/journal.pone.0225253. PMID: 31730633. PMCID: PMC6857916.

25. Avanzo M, Wei L, Stancanello J, Vallieres M, Rao A, Morin O, et al. 2020; Machine and deep learning methods for radiomics. Med Phys. 47:e185–e202. DOI: 10.1002/mp.13678. PMID: 32418336. PMCID: PMC8965689.

26. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. 2012; Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 48:441–446. DOI: 10.1016/j.ejca.2011.11.036. PMID: 22257792. PMCID: PMC4533986.

27. Liang M, Cai Z, Zhang H, Huang C, Meng Y, Zhao L, et al. 2019; Machine learning-based analysis of rectal cancer MRI radiomics for prediction of metachronous liver metastasis. Acad Radiol. 26:1495–1504. DOI: 10.1016/j.acra.2018.12.019. PMID: 30711405.

28. Yan Y, Liu H, Mao K, Zhang M, Zhou Q, Yu W, et al. 2019; Novel nomograms to predict lymph node metastasis and liver metastasis in patients with early colon carcinoma. J Transl Med. 17:193. DOI: 10.1186/s12967-019-1940-1. PMID: 31182111. PMCID: PMC6558904.

29. Li Y, Eresen A, Shangguan J, Yang J, Lu Y, Chen D, et al. 2019; Establishment of a new non-invasive imaging prediction model for liver metastasis in colon cancer. Am J Cancer Res. 9:2482–2492.
30. Lee S, Choe EK, Kim SY, Kim HS, Park KJ, Kim D. 2020; Liver imaging features by convolutional neural network to predict the metachronous liver metastasis in stage I-III colorectal cancer patients based on preoperative abdominal CT scan. BMC Bioinformatics. 21:382. DOI: 10.1186/s12859-020-03686-0. PMID: 32938394. PMCID: PMC7495853.

31. Xiao C, Zhou M, Yang X, Wang H, Tang Z, Zhou Z, et al. 2022; Accurate prediction of metachronous liver metastasis in stage I-III colorectal cancer patients using deep learning with digital pathological images. Front Oncol. 12:844067. DOI: 10.3389/fonc.2022.844067. PMID: 35433467. PMCID: PMC9010865.

32. Hao M, Li H, Wang K, Liu Y, Liang X, Ding L. 2022; Predicting metachronous liver metastasis in patients with colorectal cancer: development and assessment of a new nomogram. World J Surg Oncol. 20:80. DOI: 10.1186/s12957-022-02558-6. PMID: 35279173. PMCID: PMC8918281.

33. Chok AY, Zhao Y, Chen HLR, Tan IE, Chew DHW, Zhao Y, et al. 2023; Elderly patients over 80 years undergoing colorectal cancer resection: development and validation of a predictive nomogram for survival. World J Gastrointest Surg. 15:892–905. DOI: 10.4240/wjgs.v15.i5.892. PMID: 37342856. PMCID: PMC10277950.

34. Xiao J, Wang Y, Chen J, Xie L, Huang J. 2021; Impact of resampling methods and classification models on the imbalanced credit scoring problems. Information Sciences. 569:508–526. DOI: 10.1016/j.ins.2021.05.029.

35. International Joint Conferene on Artificial Intelligence (IJCAI). In : The 1995 International Joint Conference on AI; 1995 Aug 20-25; Montreal, Canada. Montreal: International Joint Conferene on Artificial Intelligence (IJCAI);1995. p. 1137–1143.
36. Javaid M, Haleem A, Singh RP, Suman R, Rab S. 2022; Significance of machine learning in healthcare: features, pillars and applications. Int J Intell Netw. 3:58–73. DOI: 10.1016/j.ijin.2022.05.002.

37. Alba AC, Agoritsas T, Walsh M, Hanna S, Iorio A, Devereaux P, et al. 2017; Discrimination and calibration of clinical prediction models: users' guides to the medical literature. JAMA. 318:1377–1384. DOI: 10.1001/jama.2017.12126. PMID: 29049590.

38. Schulz A, Viktil E, Godt JC, Johansen CK, Dormagen JB, Holtedahl JE, et al. 2016; Diagnostic performance of CT, MRI and PET/CT in patients with suspected colorectal liver metastases: the superiority of MRI. Acta Radiol. 57:1040–1048. DOI: 10.1177/0284185115617349. PMID: 26622057.

39. Niekel MC, Bipat S, Stoker J. 2010; Diagnostic imaging of colorectal liver metastases with CT, MR imaging, FDG PET, and/or FDG PET/CT: a meta-analysis of prospective studies including patients who have not previously undergone treatment. Radiology. 257:674–684. DOI: 10.1148/radiol.10100729. PMID: 20829538.

40. Forghani R, Savadjiev P, Chatterjee A, Muthukrishnan N, Reinhold C, Forghani B. 2019; Radiomics and artificial intelligence for biomarker and prediction model development in oncology. Comput Struct Biotechnol J. 17:995–1008. DOI: 10.1016/j.csbj.2019.07.001. PMID: 31388413. PMCID: PMC6667772.

41. Fan L, Fang M, Tu W, Zhang D, Wang Y, Zhou X, et al. 2019; Radiomics signature: a biomarker for the preoperative distant metastatic prediction of stage I nonsmall cell lung cancer. Acad Radiol. 26:1253–1261. DOI: 10.1016/j.acra.2018.11.004. PMID: 30527455.

42. Zhang L, Dong D, Li H, Tian J, Ouyang F, Mo X, et al. 2019; Development and validation of a magnetic resonance imaging-based model for the prediction of distant metastasis before initial treatment of nasopharyngeal carcinoma: a retrospective cohort study. EBioMedicine. 40:327–335. DOI: 10.1016/j.ebiom.2019.01.013. PMID: 30642750. PMCID: PMC6413336.

43. Coroller TP, Grossmann P, Hou Y, Velazquez ER, Leijenaar RT, Hermann G, et al. 2015; CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 114:345–350. DOI: 10.1016/j.radonc.2015.02.015. PMID: 25746350. PMCID: PMC4400248.

44. Busse PJ, Mathur SK. 2010; Age-related changes in immune function: effect on airway inflammation. J Allergy Clin Immunol. 126:690–699. DOI: 10.1016/j.jaci.2010.08.011. PMID: 20920759. PMCID: PMC3297963.

45. Parcesepe P, Giordano G, Laudanna C, Febbraro A, Pancione M. 2016; Cancer-associated immune resistance and evasion of immune surveillance in colorectal cancer. Gastroenterol Res Pract. 2016:6261721. DOI: 10.1155/2016/6261721. PMID: 27006653. PMCID: PMC4781955.

46. McConnell BB, Yang VW. 2009; The role of inflammation in the pathogenesis of colorectal cancer. Curr Colorectal Cancer Rep. 5:69–74. DOI: 10.1007/s11888-009-0011-z. PMID: 19756239. PMCID: PMC2743014.

47. Smith HA, Kang Y. 2013; The metastasis-promoting roles of tumor-associated immune cells. J Mol Med (Berl). 91:411–429. DOI: 10.1007/s00109-013-1021-5. PMID: 23515621. PMCID: PMC3697909.

48. Kitamura T, Qian BZ, Pollard JW. 2015; Immune cell promotion of metastasis. Nat Rev Immunol. 15:73–86. DOI: 10.1038/nri3789. PMID: 25614318. PMCID: PMC4470277.

49. Pancione M, Giordano G, Remo A, Febbraro A, Sabatino L, Manfrin E, et al. 2014; Immune escape mechanisms in colorectal cancer pathogenesis and liver metastasis. J Immunol Res. 2014:686879. DOI: 10.1155/2014/686879. PMID: 24741617. PMCID: PMC3987978.

50. Hendifar A, Yang D, Lenz F, Lurje G, Pohl A, Lenz C, et al. 2009; Gender disparities in metastatic colorectal cancer survival. Clin Cancer Res. 15:6391–6397. DOI: 10.1158/1078-0432.CCR-09-0877. PMID: 19789331. PMCID: PMC2779768.

51. Press OA, Zhang W, Gordon MA, Yang D, Lurje G, Iqbal S, et al. 2008; Gender-related survival differences associated with EGFR polymorphisms in metastatic colon cancer. Cancer Res. 68:3037–3042. DOI: 10.1158/0008-5472.CAN-07-2718. PMID: 18413774.

52. Abancens M, Bustos V, Harvey H, Mcbryan J, Harvey BJ. 2020; Sexual dimorphism in colon cancer. Front Oncol. 10:607909. DOI: 10.3389/fonc.2020.607909. PMID: 33363037. PMCID: PMC7759153.

53. Roshan MH, Tambo A, Pace NP. 2016; The role of testosterone in colorectal carcinoma: pathomechanisms and open questions. EPMA J. 7:22. DOI: 10.1186/s13167-016-0071-5. PMID: 27833666. PMCID: PMC5103431.

54. Milette S, Hashimoto M, Perrino S, Qi S, Chen M, Ham B, et al. 2019; Sexual dimorphism and the role of estrogen in the immune microenvironment of liver metastases. Nat Commun. 10:5745. DOI: 10.1038/s41467-019-13571-x. PMID: 31848339. PMCID: PMC6917725.

55. Su BB, Shi H, Wan J. 2012; Role of serum carcinoembryonic antigen in the detection of colorectal cancer before and after surgical resection. World J Gastroenterol. 18:2121–2126. DOI: 10.3748/wjg.v18.i17.2121. PMID: 22563201. PMCID: PMC3342612.

56. Hammarström S. 1999; The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Sem Cancer Biol. 9:67–81. DOI: 10.1006/scbi.1998.0119. PMID: 10202129.

57. Kamphues C, Andreatos N, Kruppa J, Buettner S, Wang J, Sasaki K, et al. 2021; The optimal cut-off values for tumor size, number of lesions, and CEA levels in patients with surgically treated colorectal cancer liver metastases: an international, multi-institutional study. J Surg Oncol. 123:939–948. DOI: 10.1002/jso.26361. PMID: 33400818.

58. Fletcher RH. 1986; Carcinoembryonic antigen. Ann Intern Med. 104:66–73. DOI: 10.7326/0003-4819-104-1-66. PMID: 3510056.

59. Duffy MJ. 2001; Carcinoembryonic antigen as a marker for colorectal cancer: is it clinically useful? Clin Chem. 47:624–630. DOI: 10.1093/clinchem/47.4.624. PMID: 11274010.

60. Hall C, Clarke L, Pal A, Buchwald P, Eglinton T, Wakeman C, et al. 2019; A review of the role of carcinoembryonic antigen in clinical practice. Ann Coloproctol. 35:294–305. DOI: 10.3393/ac.2019.11.13. PMID: 31937069. PMCID: PMC6968721.

61. Enquist IB, Good Z, Jubb AM, Fuh G, Wang X, Junttila MR, et al. 2014; Lymph node-independent liver metastasis in a model of metastatic colorectal cancer. Nat Commun. 5:3530. DOI: 10.1038/ncomms4530. PMID: 24667486.

62. Ahana P, Kavitha G. 2022; Radiomic features based severity prediction in dementia MR images using hybrid SSA-PSO optimizer and multi-class SVM classifier. IRBM. 43:549–560. DOI: 10.1016/j.irbm.2022.05.003.

63. Awad M, Khanna R. Awad M, Khanna R, editors. 2015. Support vector machines for classification. Efficient Learning Machines: Theories, Concepts, and Applications for Engineers and System Designers. p. 39–66. Apress;DOI: 10.1007/978-1-4302-5990-9_3.

64. Prasad AM, Iverson LR, Liaw A. 2006; Newer classification and regression tree techniques: bagging and random forests for ecological prediction. Ecosystems. 9:181–199. DOI: 10.1007/s10021-005-0054-1.

Fig. 1
Tree diagram of predictive modeling algorithms included in the systematic review. LASSO, least absolute shrinkage and selection operator.
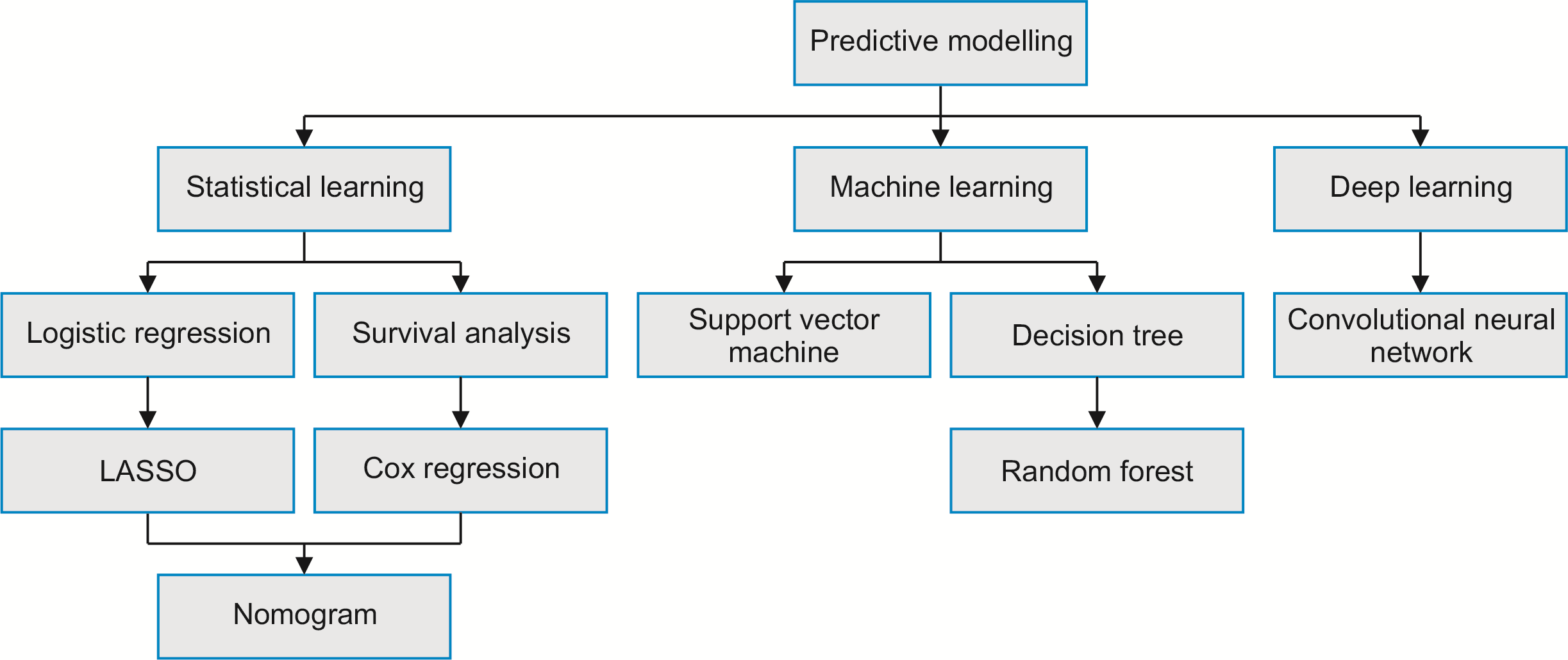
Fig. 2
PRISMA flow diagram for data collection. The search returned a total of 141 records, of which 7 studies that reported predictive modeling techniques to predict colorectal caner liver metastasis (CRCLM) were included in the systematic review.
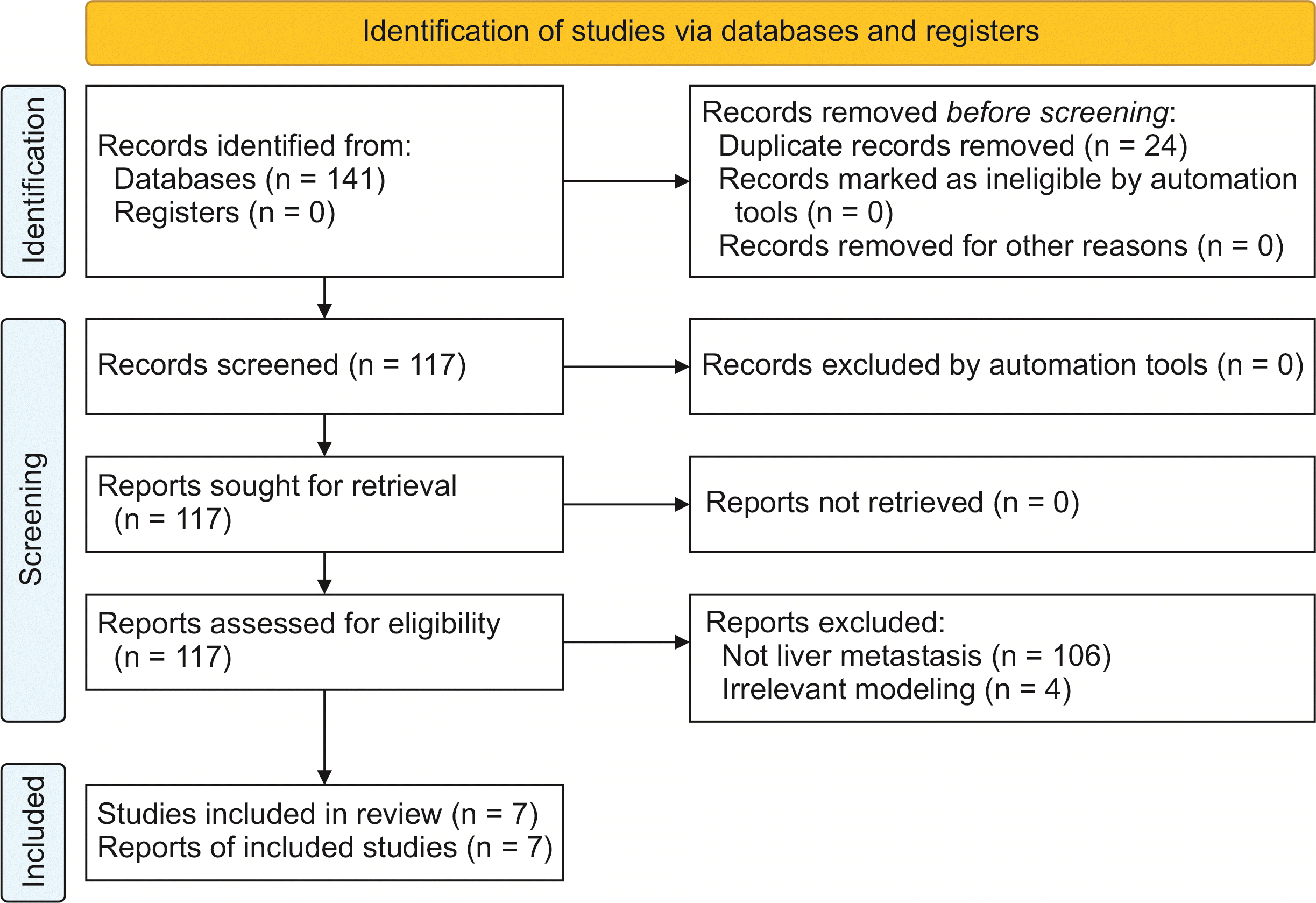
Fig. 3
Methodological evaluation of the included predictive models. Assessment of the RoB based on PROBAST criteria. (A) Summary of RoB assessment. (B) Summary of applicability assessment. PROBAST, prediction model risk of bias assessment tool; RoB, risk of bias.
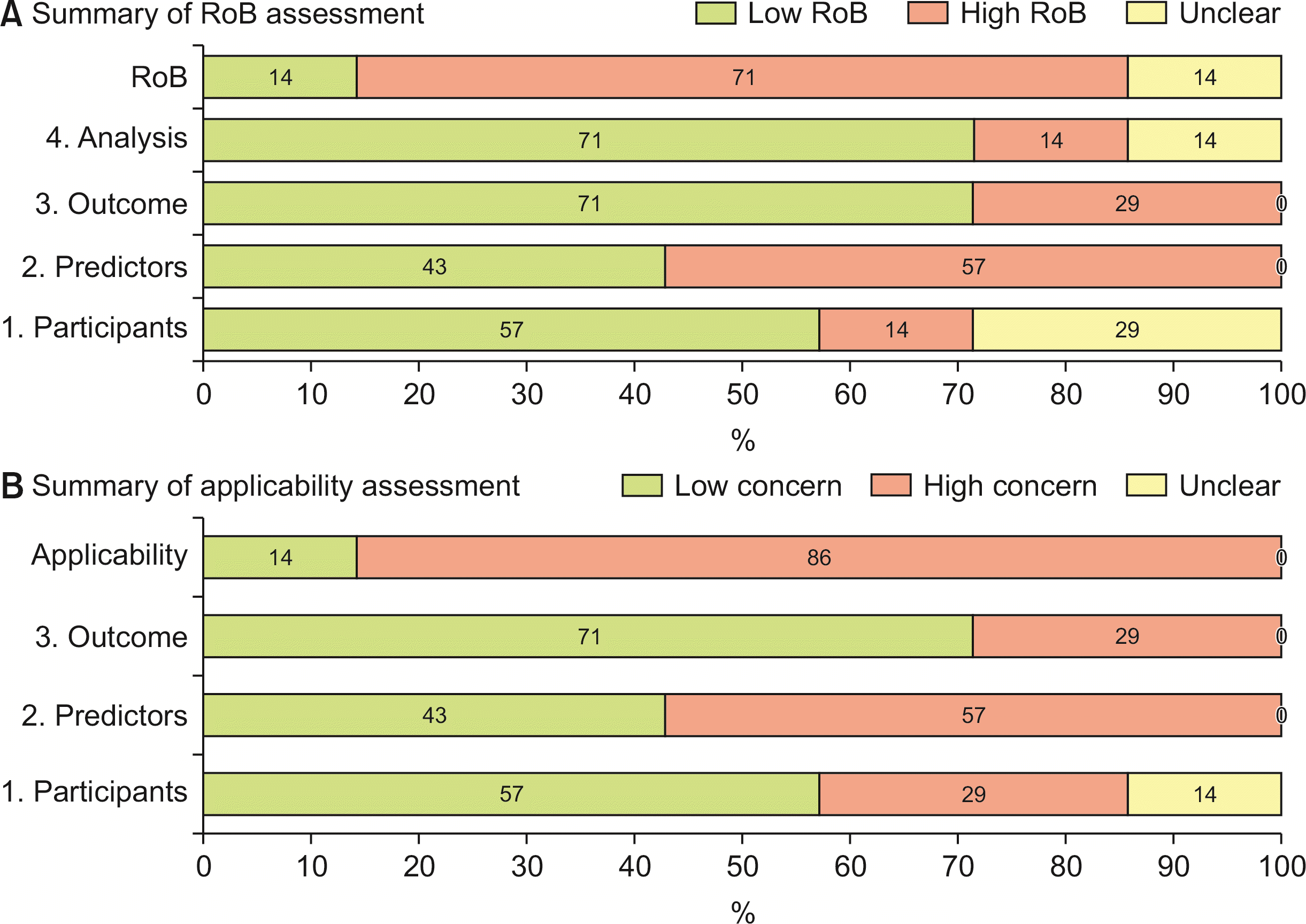
Table 1
Predictive modeling terminology included in the systematic review
Table 2
Characteristics of included studies
| Author | Year | Country | Study design | Study period | Study setting | Disease condition | Surgical procedure | Sample size | Predictive model | Internal validation | External validation | Discrimination | Calibration | Predictor |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liang et al. [27] | 2019 | China | Retrospective | 2011–2017 | Single-center | Rectal cancer | Total mesorectal excision | 108 | Radiomics + LR | Cross-validation | No | AUC = 0.740 | No | 22 radiomic features |
| Radiomics + SVM | AUC = 0.770 | |||||||||||||
| Yan et al. [28] | 2019 | China | Retrospective | 2004–2015 | Single-center | Colon carcinoma | Not specified | 32,819 | Cox regression; Nomogram | Bootstrapping | No | AUC = 0.825 | Yes | Age; CEA; tumor size; tumor grade; N staging |
| Li et al. [29] | 2019 | China | Retrospective | 2015–2018 | Single-center | Colon cancer | Radical colectomy | 48 | Clinical + SVM | Cross-validation | No | AUC = 0.690 | No | Age; sex; tumor location; tumor histology; tumor size |
| Radiomics + SVM | AUC = 0.850 | 6 radiomic features | ||||||||||||
| Hybrid + SVM | AUC = 0.870 | 6 clinical and radiomic features | ||||||||||||
| Taghavi et al. [15] | 2021 | Netherlands | Retrospective | 2006–2016 | Multicenter | Colorectal cancer | Not specified | 91 | Clinical + RF | Bootstrapping; Cross-validation | No | AUC = 0.860 | No | Age; sex (male/female); primary tumor site; tumor stage; nodal stage; CEA; chemotherapy |
| Radiomics + RF | AUC = 0.710 | 101 radiomic features | ||||||||||||
| Combined + RF | AUC = 0.860 | 104 clinical and radiomic features | ||||||||||||
| Lee et al. [30] | 2020 | South Korea | Retrospective | 2008–2013 | Single-center | Colorectal cancer | Colectomy | 2,019 | CNN + LR | Cross-validation | No | AUC = 0.747 | No | Age; sex (male/female); T stage; N stage; CT image features |
| CNN + RF | AUC = 0.697 | |||||||||||||
| Xiao et al. [31] | 2022 | China | Retrospective | 2016–2017 | Single-center | Colorectal cancer | Radical colorectal resection | 611 | CNN | Cross-validation | No | AUC = 0.758 | Yes | Digital pathological images |
| Cox regression; Nomogram | Bootstrapping | AUC = 0.848 | Digital pathological images; N stage; T stage; VE/LI/PI | |||||||||||
| Hao et al. [32] | 2022 | China | Retrospective | 2016–2019 | Single-center | Colorectal cancer | Not specified | 293 | LR; Nomogram | Bootstrapping | No | AUC = 0.784 | Yes | Age; CEA; VI; T stage; N stage; family CRCLM history; KRAS |
AUC, area under the receiver operating characteristic curve; CEA, carcinoembryonic antigen; CNN, convolutional neural network; CRCLM, colorectal cancer liver metastasis; KRAS, Kirsten rat sarcoma viral oncogene homologue; LI, lymphatic invasion; LR, logistic regression; PI, perineurial invasion; RF, random forest; SVM, support vector machine; VE, vascular emboli; VI, vascular invasion.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download