Abstract
Background: Ideal dose of the antimicrobials should be decided by considering their killing dynamics since sufficient elimination of the causative microorganisms is critical for proper antimicrobial treatment. In this study, the bactericidal activities of carbapenems by resistance mechanisms were assessed for carbapenem-resistant Pseudomonas aeruginosa. Methods: Minimal inhibition concentrations (MICs) of carbapenems were determined by broth dilution method and the resistance mechanisms were identified by PCR and DNA sequencing. The expression levels of efflux pumps were determined by reverse transcriptase real-time PCR. Time-kill curves were plotted by time-course numeration of the viable cells grown under imipenem and meropenem at 1× and 4× MICs, respectively. Results: One P. aeruginosa strain was susceptible, whereas three were resistant to carbapenems by defective OprD, efflux pump overproduction, and/or IMP-6 production. The susceptible strain had imipenem and meropenem MICs of 2 and 1 mg/L, respectively. The MICs were elevated by eight-fold by defective OprD, 16- and 32-fold by the pump overproduction, and four- and <64-fold by the combination of two determinants and the IMP-6 carbapenemase. While both the carbapenems showed time-dependent bactericidal activity to the susceptible isolate, either of the carbapenem-resistant determinants, such as decreased membrane permeability, carbapenemase production, or the defective OprD,presented concentration-dependent bacteriostatic activity.Conclusion: Different killing dynamics of the carbapenems were observed depending on the resistance determinants, and the results would guide a proper treatment strategy for the patients using these drugs.
Pseudomonas aeruginosa is an opportunistic pathogen causative for healthcare-associated infections [1]. Carbapenems are a major treatment option for infections caused by drug-resistant P. aeruginosa. However, increasing proportion of the carbapenem resistance in the P. aeruginosa clinical strains is of concern [2]. The resistance mechanism of the drug includes i) membrane impermeability either by deficiency of the outer-membrane porin OprD or by overexpression of the resistance-nodulation cell division (RND) efflux pumps such as MexAB-OprM and MexXY-OprM [3] and ii) acquisition of the carbapenemase gene, such as a class A Guiana extended-spectrum beta-lactamase (GES) of some subtypes and class B imipenemase (IMP) and Verona integron-encoded metallo-beta-lactamase (MBLs) (VIM) [4]. The level of resistance depends on the resistance determinants. The carbapenemase confers mid- to high-level resistance, while membrane impermeability leads only reduced susceptibility. The clinical strains often carry two or more resistance determinants and the interplay of multiple determinants has been demonstrated to confer higher level resistance to carbapenems [5].
Since elimination of the infection-causative microorganisms is critical to succeed the treatment, antimicrobial doses should be decided in consideration of the killing dynamics. Consequently, characterizing the killing effect of antimicrobials, including its bacteriostatic and bactericidal activity, is principal to optimize antimicrobial pharmacodynamics. While many efforts have been made to know the killing dynamics of drugs against drug-susceptible bacteria [6], that against the drug-resistant bacteria harboring single or multiple resistance determinants to carbapenems has been insufficiently established and the altered killing dynamics by common resistance determinant is needed. And in this study, we evaluated the killing dynamics of carbapenems against P. aeruginosa strains having varied common mechanisms of carbapenem resistance.
Bacterial strains and Antimicrobial susceptibility testing
Four P. aeruginosa blood isolates from three general hospitals in South Korea between 2016 and 2017 were used. Bacterial species were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry using a Bruker MALDI Biotyper® instrument (Bruker, Billerica, MA, USA) under the criteria of the log score above 2.0. Minimal inhibitory concentrations (MICs) of imipenem (Sigma-Aldrich, St. Louis, MO, USA) and meropenem (Sigma-Aldrich) were determined by broth microdilution methods using Mueller-Hinton (MH) broth and both Escherihia coli ATCC 25922 and P. aeruginosa ATCC 27853 were used for quality control [7].
DNA manipulation and polymerase chain reaction (PCR)
Total DNA was extracted using MG Genomic DNA Purification kit (MG MED Inc., Seoul, Korea). PCR and following Sanger sequencing was carried out for known acquired carbapenemases, such as blaIMP, blaVIM, blaKPC, blaNDM, and blaGES [8], and the oprD gene [9].
Determination of the efflux pump overexpression
Total RNA was extracted from exponentially growing bacterial cells by using RNeasy® plus mini (Qiagen, Hilden, Gemany). Levels of mRNA for the three RND efflux pumps, and the house-keeping rpoB gene was quantified by LightCycler® 480 instrument II (Roche Diagnostics, Rotkreuz, Switzerland) using the LightCycler® RNA amplification kit with SYBR green I (Roche Diagnostics) using gene-specific primer pairs 5ʹ-CAAGGGCGTCGGTGACTTCCA-3ʹ and 5ʹ-ACCTGGGAACCGTCGGGATTGA-3ʹ for mexB, 5ʹ-GGACCACGCCGAAACCGAACG-3ʹ and 5ʹ-CGCCGCAACTGACCCGCTACA-3ʹ for mexY, 5ʹ-GGACGGCTCGCTGGTCCGGCT-3ʹ and 5ʹ-CGACGAAGCGCGAGGTGTCGT-3ʹ for mexD, 5ʹ-CTTGGTACGACCGTTCACGT-3ʹ and 5ʹ-GCTGAAACTGAACCACCTGG-3ʹ for rpoB with the following cycles: predenaturtation at 95°C for 30 s, followed by 45 cycles at 95°C for 5 s, 58°C for 10 s, and 72°C for 20 s. The transcriptome levels of each pump were normalized to that of rpoB, and relative levels were calculated by dividing those of the P. aeruginosa A1701DS. Each experiment was performed in duplicate twice independently.
Time–kill assay
Each strain was grown in MH broth till mid-exponential phase and were diluted to have 106 colony forming units (CFU)/ml. Drugs were then added at 1× (2 mg/L imipenem and 1 mg/L meropenem) and 4× MICs (8 mg/L imipenem and 4 mg/L meropenem). CFUs were numerated by plating every two hours and plotted. The killing kinetics were determined twice independently, and verified the results being reproducible. Bactericidal activity was defined as a ≥3.0 log CFU/ml decrease in bacterial count at any time point during the 24-hour exposure of the drug.
Carbapenem susceptibility and the relevant mechanism
The four P. aeruginosa strains studied included one carbapenem-susceptible and three carbapenem-resistant strains (Table 1). P. aeruginosa A1701DS was susceptible to both carbapenems and MICs of imipenem and meropenem were 2 mg/L and 1 mg/L, respectively. The three carbapenem-resistant strains had varied MICs of both drugs. P. aeruginosa B1707∆oprD strain had the imipenem MIC of 16 mg/L and the meropenem MIC of 8 mg/L. P. aeruginosa F1709pump↑ had both carbapenem MICs of 32 mg/L, and B1605∆oprD/ pump↑/IMP had MICs of 8 mg/L and >64 mg/L of imipenem and meropenem, respectively.
Except the carbapenem-susceptible P. aeruginosa A1701DS, the three P. aeruginosa strains had single or multiple mechanisms of carbapenem resistance (Table 1). Defective OprD was identified in P. aeruginosa B1707∆oprD and B1605∆oprD/ pump↑/IMP strains. Overexpression of the RND pump was observed in two P. aeruginosa strains. The MexAB-OprM RND efflux pump of the three tested RND efflux systems was 1.40- fold overexpressed in P. aeruginosa F1709pump↑. For the P. aeruginosa B1605∆oprD/ pump↑/IMP, 1.64-fold overexpression of the MexAB-OprM, 3.37-fold overexpression of the MexXY-OprM, and 2.84-fold overexpression of the MexCD-OprJ RND efflux system were observed. The P. aeruginosa B1605∆oprD/ pump↑/IMP harbored the blaIMP-6 gene for MBL.
Time-kill kinetics of carbapenems on P. aeruginosa carrying varied mechanisms of resistance
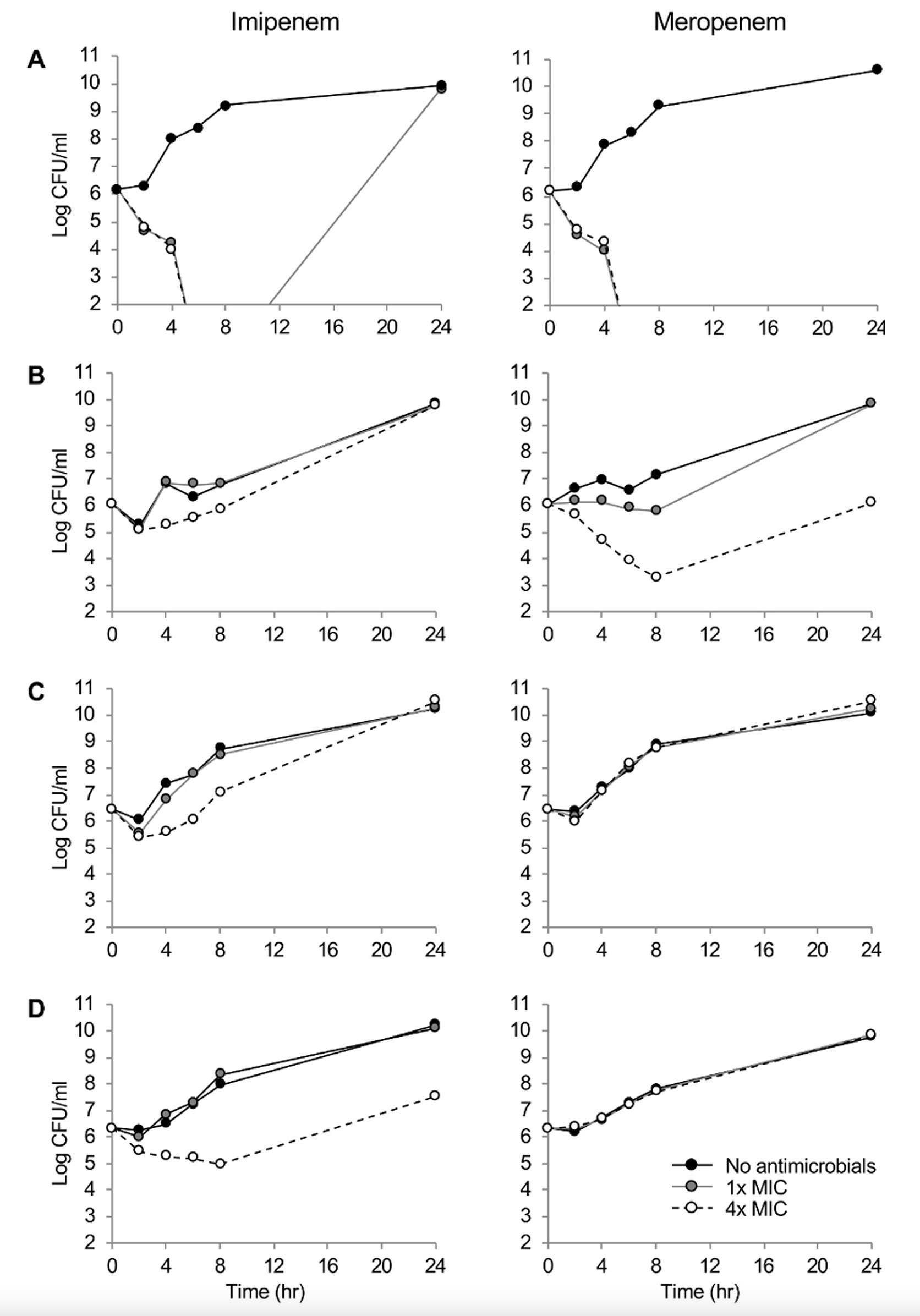
Fig. 1 showed the killing dynamics of imipenem and meropenem. Both carbapenems demonstrated rapid time-dependent killing kinetics against the drug-susceptible P. aeruginosa A1701DS strain at 1×and 4×MICs. By the 24-hour incubation at 1×MIC of imipenem, regrowth of the strain was observed. The meropenem presented clear concentration-dependent killing dynamics against P. aeruginosa B1707∆oprD. Within 8 hours, elimination of 3.0 log CFU/ml were observed. The imipenem presented concentration-dependent killing kinetics against P. aeruginosa B1707∆oprD, however, it only exhibted 1.0 log CFU/ml elimination by 2 hours. Finally, both the RND pump-overproducing strains, the F1709pump↑ and the B1605∆oprD/ pump↑/IMP were not killed by meropenem at all, while concentration-dependent killing dynamics by imipenem were observed.
Adequate antimicrobial therapy is critical for the outcome of the patients with infectious diseases and the in vitro kinetics of bacterial killing is one of the key factors for guiding the proper antimicrobial choice as dead bugs don’t mutate [10]. Even though the definitions of “bacteriostatic” and “bactericidal” are clear, no antimicrobial agents could be definitively categorized into the two, since the killing kinetics of the drug is dependent on the targeting bacteria, inoculum size, and so on [11].
The carbapenems are one of the last-line drugs to treat P. aeruginosa infections. Since the drugs are used empirically for the critically ill patients, resistance to the drug is critical for the patient’s outcome [12]. The three resistance determinants, i.e. OprD deficiency, overproduction of the RND efflux pump, and MBL production, are common mechanisms of resistance to carbapenems globally in P. aeruginosa [13]. In addition, the IMP-6-producing P. aeruginosa used in the study is disseminated in South Korea since the end of the 2000s [9,14]. Thus, bacterial killing effects of carbapenems against the P. aeruginosa carrying the resistance determinants was needed to be evaluated.
Carbapenems showed a time-dependent bactericidal activity to the susceptible isolate as the most beta-lactams do. Certain regrowth of the bacteria by 24 hours under the 1×MIC imipenem was likely due to the collaborative effect of the poor chemical stability of the drug and the intrinsic cephalosporinase leaky in P. aeruginosa, which has a very weak carbapenemase-hydrolyzing activity. While deficiency of the OprD elevated the MICs of both carbapenems equally by eight folds, clear concentration-dependent killing kinetics of the meropenem was observed against the OprD-defective strain by eliminating 3.0 log CFU/ml in eight hours at the 4×MIC. Conceivably, the difference was led by imipenem preference of the porin [15]. Meropenem MICs were much more elevated by overproduction of the efflux pump, especially that of the MexAB-OprM having preference to meropenem [3,16]. The IMP-6 carbapenemase production resulted in additional elevation of the meropenem MIC on top of the RND efflux pump overexpression. Imipenem susceptibility was not affected by the IMP-6 making a good correlation with the previous report of the enzyme hydrolyzing efficiently the meropenem rather than the imipenem [17]. Further attention is needed to the carbapenemase producers, since they often harbor class 1 integron including multiple gene cassettes of drug resistance [18] resulting in multidrug-resistance elevating mortality of the victimized patients [19].
Notably, the mechanisms of carbapenem resistance threat the patients not only by limiting the drug choices for treatment, but also by putting the patients at risk through its alternative role as a virulence factor, and the point highlights the importance to identify the actual mechanism of carbapenem resistance. Recently, the OprD deficiency in P. aeruginosa was recognized as a risk factor for the patients with P. aeruginosa bloodstream infections by augmenting the 6-week mortality from 17.2% to 33.3% [20]. The RND efflux pump is also known to be associated with the bacterial virulence mostly by involving in the biofilm formation and quorum sensing [21,22].
This study has an obvious imperfection of limited number of strains. However, the killing kinetics of carbapenems would help to better optimize the antimicrobial pharmacodynamics against P. aeruginosa clinical strains harboring common mechanisms of carbapenem resistance. To conclude, both the time-dependent killing kinetics and bactericidal activity of carbapenems were affected by any of the common resistance determinants. The defective OprD diminished the killing activity of imipenem rather than that of meropenem, while the RND pump overproduction and the meropenemase MBL IMP-6 weaken the activity of meropenem than that of imipenem.
배경: 최적의 항균제 투여에 의한 감염 원인균의 효율적 제균은 세균 감염증 치료에 필수적이며, 이상적인 항균제 투여량은 약물의 살균 동역학을 바탕으로 결정된다. 본 연구에서는 다양한 내성 기전의 카바페넴 내성 녹농균에 대한 카바페넴의 살균력을 평가하였다.
방법: 환자의 혈액검체에서 분리된 녹농균을 사용하였다. 카바페넴의 최소 억제농도는 미량 액체배지 희석법으로 결정하였고, 카바페넴 내성 결정요소는 연쇄중합효소반응 및 염기서열 분석을 통하여 규명하였다. 항생제 유출펌프의 발현 정도는 실시간 역전사 연쇄중합효소반응으로 측정하였다. 최소 억제농도의 1배 또는 4배 농도의 이미페넴이나 메로페넴 용액에서 녹농균 균주를 배양하며 매 시간 집락 수를 측정하여 살균 동역학 커브를 작성하였다.
결과: 실험 대상 균주 4주 중 1주는 카바페넴에 감수성이었으며, 3주는 OprD 결손, 항생제 유출펌프 과생성 및 IMP-6 효소 생성으로 인해 카바페넴 내성을 획득한 균주였다. 감수성 균주의 이미페넴 최소 억제농도는 2 mg/L, 메로페넴은 1 mg/L 이었으며, 이들 두개 항생제의 최소 억제농도는 OprD 결손에 의해 각 8배, 항생제 유출펌프 과발현에 의해 16 및 32배, OprD 결손, 항생제 유출펌프 과발현, IMP-6 생성의 3가지 내성결정요소에 의해 각 4배와 >64배 상승하였다. 카바페넴 감수성 균주는 이미페넴과 메로페넴 모두에 대해 시간 의존성 살균효과를 나타낸 반면, 카바페넴 내성 균주는 농도 의존성 정균효과를 보였다. 이미페넴은 OprD 결손 또는 IMP-6 생성 균주, 메로페넴은 OprD 결손균주에 대하여 농도 의존성 정균효과가 있었다.
결론: 카바페넴은 녹농균의 내성 기전에 따라 상이한 살균 동역학을 보였다. 본 연구의 결과는 카바페넴 내성 녹농균 감염증의 항균제 치료 가이드라인 확립에 도움을 줄 것으로 생각한다.
FUNDING
This research was supported by a fund (NRF-2018R1C1B6002674) from National Research Foundation of Korea.
REFERENCES
1. Poole K. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2011;2:65.
2. Lee H, Yoon EJ, Kim D, Jeong SH, Won EJ, Shin JH, et al. Antimicrobial resistance of major clinical pathogens in South Korea, May 2016 to April 2017: first one-year report from KorGLASS. Euro Surveill 2018;23:1800047.
3. Livermore DM. Of Pseudomonas, porins, pumps and carbapenems. J Antimicrob Chemother 2001;47:247-50.
4. Hong DJ, Bae IK, Jang IH, Jeong SH, Kang HK, Lee K. Epidemiology and characteristics of metallo-beta-lactamase-producing Pseudomonas aeruginosa. Infect Chemother 2015;47:81-97. .
5. Quale J, Bratu S, Gupta J, Landman D. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 2006;50:1633-41.
6. Mattoes HM, Kuti JL, Drusano GL, Nicolau DP. Optimizing antimicrobial pharmacodynamics: dosage strategies for meropenem. Clin Ther 2004;26:1187-98.
7. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. CLSI Document M100S. 26th ed. Wayne; PA: 2016.
8. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011;17:1791-8.
9. Hong JS, Yoon EJ, Lee H, Jeong SH, Lee K. Clonal dissemination of Pseudomonas aeruginosa sequence type 235 isolates carrying blaIMP-6 and emergence of blaGES-24 and blaIMP-10 on novel genomic islands PAGI-15 and -16 in South Korea. Antimicrob Agents Chemother 2016;60:7216-23.
10. Stratton CW. Dead bugs don't mutate: susceptibility issues in the emergence of bacterial resistance. Emerg Infect Dis 2003;9:10-6.
11. Pankey GA and Sabath LD. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of gram-positive bacterial infections. Clin Infect Dis 2004;38:864-70.
12. Dantas RC, Ferreira ML, Gontijo-Filho PP, Ribas RM. Pseudomonas aeruginosa bacteraemia: independent risk factors for mortality and impact of resistance on outcome. J Med Microbiol 2014;63:1679-87.
13. Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009-11 in14 European and Mediterranean countries. J Antimicrob Chemother 2014;69:1804-14.
14. Seok Y, Bae IK, Jeong SH, Kim SH, Lee H, Lee K. Dissemination of IMP-6 metallo-betalactamase-producing Pseudomonas aeruginosa sequence type 235 in Korea. J Antimicrob Chemother 2011;66:2791-6.
15. Huang H and Hancock RE. Genetic definition of the substrate selectivity of outer membrane porin protein OprD of Pseudomonas aeruginosa. J Bacteriol 1993;175:7793-800.
16. Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2000;44:3322-7.
17. Yano H, Kuga A, Okamoto R, Kitasato H, Kobayashi T, Inoue M. Plasmid-encoded metallobeta-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob Agents Chemother 2001;45:1343-8.
18. Nordmann P, Naas T, Fortineau N, Poirel L. Superbugs in the coming new decade; multidrug resistance and prospects for treatment of Staphylococcus aureus, Enterococcus spp. and Pseudomonas aeruginosa in 2010. Curr Opin Microbiol 2007;10:436-40.
19. Woodford N, Turton JF, Livermore DM. Multiresistant gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 2011;35:736-55.
20. Yoon EJ, Kim D, Lee H, Lee HS, Shin JH, Park YS, et al. Mortality dynamics of Pseudomonas aeruginosa bloodstream infections and the influence of defective OprD on mortality: prospective observational study. J Antimicrob Chemother 2019;74:2774-83.
21. Alcalde-Rico M, Hernando-Amado S, Blanco P, Martinez JL. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front Microbiol 2016;7:1483.
22. Yoon EJ, Balloy V, Fiette L, Chignard M, Courvalin P, Grillot-Courvalin C. Contribution of the Ade resistance-nodulation-cell division-type efflux pumps to fitness and pathogenesis of Acinetobacter baumannii. mBio 2016;7:e00697-16.
Table 1
Carbapenem susceptibility and relative level of efflux pump gene expression in P. aeroginosa strains
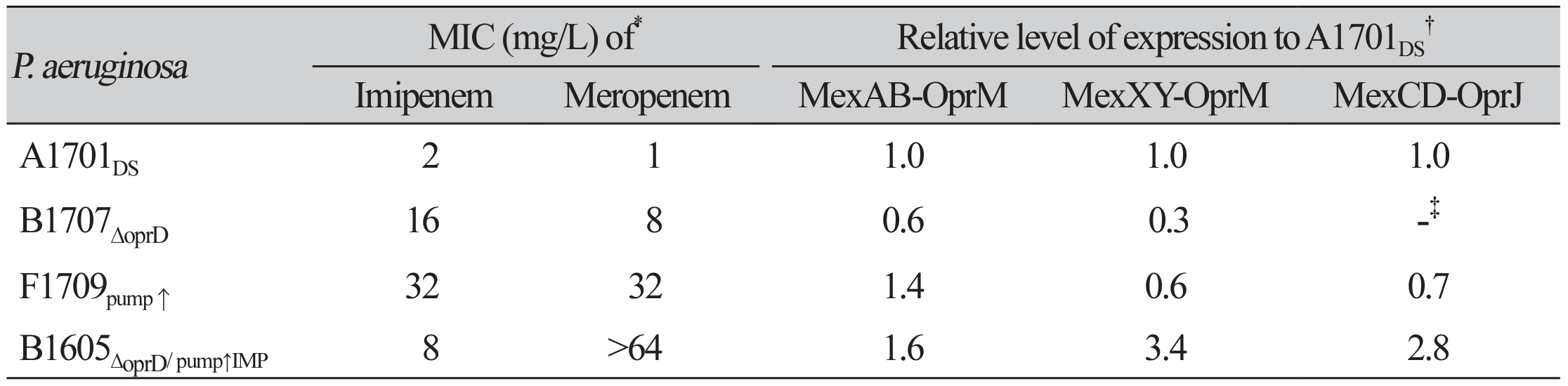
*MICs were determined by broth dilution method. Imipenem and meropenem MICs in quality control strain P. aeruginosa ATCC 27853 were 4 and 0.5 mg/L, respectively.†The relative level of pump expression was determined for the RND pump genes, mexB for MexAB-OprM, mexY for MexXY-OprM, and mexD for MexCD-OprJ.‡No mexD gene was identified by PCR.Abbreviation: MICs, minimal inhibitory concentrations.
Fig. 1.
Time–kill curves of imipenem and meropenem against P. aeruginosa (A) A1701DS, (B) B1707∆oprD, (C) F1709pump↑, (D) B1605∆oprD/pump↑/IMP strains. Number of bacterial colony forming units are presented to the detection limit (102 CFU/mL). Black circle and black solid line, no antibiotics treated; gray circle and gray solid line, 1×MIC; and open circle and black broken line, 4×MIC.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download