Abstract
Purpose
The gene expression test (GET) was used to predict the response to chemotherapy and the recurrence risk. Several randomized clinical trials have demonstrated that some patients with node-positive disease can achieve favorable survival outcomes even without adjuvant chemotherapy. This study aimed to predict the results of Oncotype DX (Genomic Health) and MammaPrint (Agendia) using traditional clinicopathological factors.
Methods
We reviewed the records of 311 patients who underwent GET for hormone receptor-positive/human epidermal growth factor receptor 2 (HER2)-negative primary invasive breast cancer with node-positive disease between 2015 and 2022 at Severance Hospital and Gangneung Asan Medical Center. Univariate and multivariate logistic regression analyses assessed the relationships between clinicopathological variables and risk stratification using the GET results.
Results
A simple scoring system was created by assigning integer values to each variable. A score of 3 was assigned for histological grade 3, a score of 2 for pathologic T2 or above, and a score of 1 for a lower progesterone receptor (1–20 or Alled score 3–6), HER2 2-positive, and high Ki-67 (>20). In the validation cohort, overall accuracy was 0.798 (95% confidence interval, 0.744–0.844).
Conclusion
The high GET risk results can be predicted using traditional clinicopathological factors: tumor size, progesterone receptor, histological grade, HER2, and Ki-67. These results will be useful for treatment decision-making among clinically high-risk patients with HR-positive/HER2-negative and node-positive disease, helping to identify patients to whom the GET assay may not apply.
Postoperative adjuvant systemic treatment for breast cancer is conducted considering various oncological factors, including age, tumor burden, hormone receptor (HR) status (estrogen receptor [ER] or progesterone receptor [PR]), human epidermal growth factor receptor 2 (HER2), and the Ki-67 index [12345]. Axillary lymph node (ALN) metastases have served as indicators of poor prognosis and criteria for adjuvant systemic treatment [6]. Even with confirmed ALN metastases, some patients with HR (+) and HER2 (−) breast cancer tend to show relatively good oncological outcomes compared with other subtypes, even without undergoing chemotherapy (CTx) [78]. Previously, CTx was uniformly administered based on anatomical staging [2]. However, due to the high survival rates in these groups, there has been a shift in research focus toward minimizing the side effects of treatments [9]. Current research aims to tailor these treatments to individuals to reduce side effects and maintain similar or noninferior survival outcomes. Among such research, the RxPONDER (Rx for Positive Node, Endocrine Responsive Breast Cancer) and MINDACT (Microarray In Node-Negative and 1 to 3 Positive Lymph Node Disease May Avoid Chemotherapy) trials are the most representative studies [78].
The Oncotype DX (ODX; Genomic Health) is a diagnostic tool that analyzes the expression of 16 breast cancer-related genes and 5 reference genes in tumor tissues, providing a recurrence score (RS) ranging from 0 to 100. This score is used to predict the future risk of recurrence and the potential benefits of CTx. MammaPrint (MMP; Agendia) analyzes 70 genes to evaluate the risk of distant metastasis within 10 years in early-stage breast cancer. This result is classified as low or high genetic risk. In the case of the low-risk group, they can expect a good prognosis without CTx, even if they are classified as clinically high risk due to ALN metastasis.
Recently, randomized controlled trials have been published using ODX and MMP in patients with breast cancer with ALN metastases. As a result of these studies, it has been demonstrated that CTx can be omitted in certain patients with HR (+), HER2 (−), and ALN metastases who are classified as clinically high risk. Based on the results of the randomized controlled trial, these gene expression tests (GETs) have become standard tools in treatment decision-making and are included in the clinical guidelines of both the American Society of Clinical Oncology and the National Comprehensive Cancer Network [10111213].
GET is an important part of decision-making in breast cancer treatment; however, several disadvantages exist. In the Korean healthcare environment, national health insurance does not cover the cost of these tests, which adds a financial burden to patients already bearing the cost of breast cancer treatment. Additionally, the time-consuming process of these tests can delay the initiation of systemic adjuvant CTx for patients until these results become available. GET is an essential tool to progress toward tailored treatment by identifying patients who could potentially forego CTx. However, the high cost and the potential for delay of CTx are major barriers. For these reasons, utilizing clinicopathological values to predict the outcomes of gene expression assays could prove beneficial. This study aimed to develop and validate a model for predicting the outcomes of ODX and MMP in patients with ER (+), HER2 (−), and ALN metastases.
The project was reviewed and approved by the Institutional Review Board (IRB) of the Severance Hospital (No. 4-2023-0725) and Gangneung Asan Medical Center (No. 2023-08-006). The patient’s consent to participate was waived by the IRB owing to the retrospective nature of the study.
Patients with primary breast cancer who underwent upfront surgery between January 2015 and December 2022 were retrospectively selected from the medical databases of Severance Hospital (Seoul, Korea) and Gangneung Asan Medical Center (Gangneung, Korea). Among these subjects, HR (+), HER2 (–), and pathologically node-positive patients were included. Inclusion criteria were refined for patients who underwent genomic profiling tests such as ODX and MMP. A total of 311 patients were included in this retrospective study to predict GET results using traditional clinicopathological factors. The medical database cataloged patient characteristics, including age at diagnosis, body mass index (BMI), and menopausal status. Additionally, it documented the type and results of GET. Furthermore, it incorporated postoperative pathological results, encompassing elements such as histological type, histological grade (HG), the size of invasive breast cancer, the number of lymph node metastases, the ER expression, PR expression, HER2 expression, and the Ki-67 index. HG was assessed using the modified Bloom-Richardson grading system. Based on the policy of each institution, tumors were classified as positive for ER and PR if they demonstrated ≥1% of nuclear-stained cells or had a score of 3 or more according to the Allred scoring method. ODX risk groups were stratified into low risk, with RSs ranging from 0 to 25, and high risk, with RSs ranging from 26 to 100. The risk groups for MMP were determined based on the results of the MMP test reports.
GET results were classified as low or high risk. In cases where clinical pathological findings are continuous variables, they are converted into binary or multinomial variables for analysis based on medical evidence or distribution. For binary or multinomial variables, according to the results of GET, a chi-square test or Fisher exact test was performed for comparison. We reviewed the patient count annually and divided the data into 2 segments: from 2015 to the year when the cumulative number of patients reached 75% and the period after that. A predictive model was developed using 75% of the cumulative patient data, and the remaining 25% was utilized for validation. Within the development cohort, linear regression was performed to investigate patient characteristics and traditional clinical pathological indicators related to high-risk groups. To make it simple and practical, points were assigned to each variable as rounded-off integer values based on the estimation values from the multivariate analysis. The simplified scoring system was validated both internally and externally with 2 cohorts. We analyzed the receiver operating characteristic curve and determined the area under the curve (AUC). All statistical analyses were conducted using SAS software ver. 9.3 (SAS Institute) and R software ver. 3.1.1 (The R Foundation), with P-values less than 0.05 deemed significant.
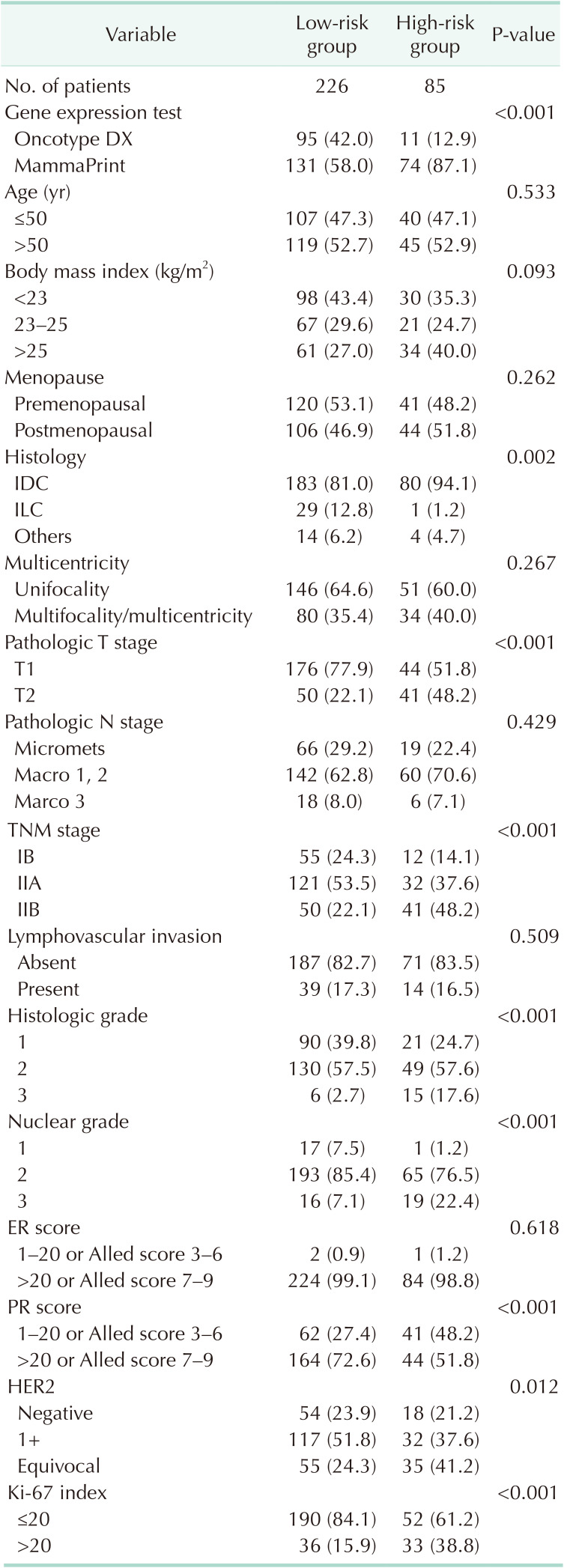
The risk classification was based on an ODX RS of 25 and the reported results for MMP. Characteristics according to the GET results are shown in Table 1. No significant differences exist between the 2 groups when categorized based on an age threshold of 50 years and menopausal status (52.3 ± 11.1 years vs. 51.73 ± 9.1 years, P = 0.658). A higher rate of subjects with a BMI of 25 kg/m2 or above was observed in the high-risk group. The histologic grade (17.6% vs. 2.7%, P < 0.001) and nuclear grade (NG; 22.4% vs. 7.1%, P < 0.001) were higher, and the size of invasive breast cancer was larger (2.0 ± 0.8 cm vs. 1.6 ± 0.6 cm, P < 0.001) in the high-risk group. However, there were no differences between the 2 groups regarding axillary nodal burden, lymphovascular invasion, or multicentricity. The ER score was predominantly high in both groups, with no significant differences observed. However, the high-risk group had a greater proportion of individuals with low PR expression (48.2% vs. 27.4%, P < 0.001), HER2 2 (+) status (41.2% vs. 24.3%, P = 0.012), and high Ki-67 (38.8% vs. 15.9%, P < 0.001).
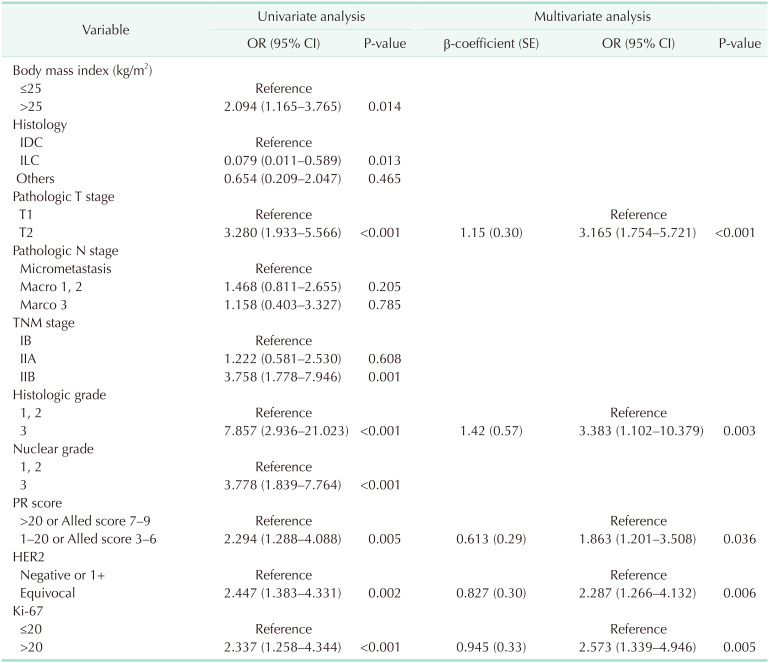
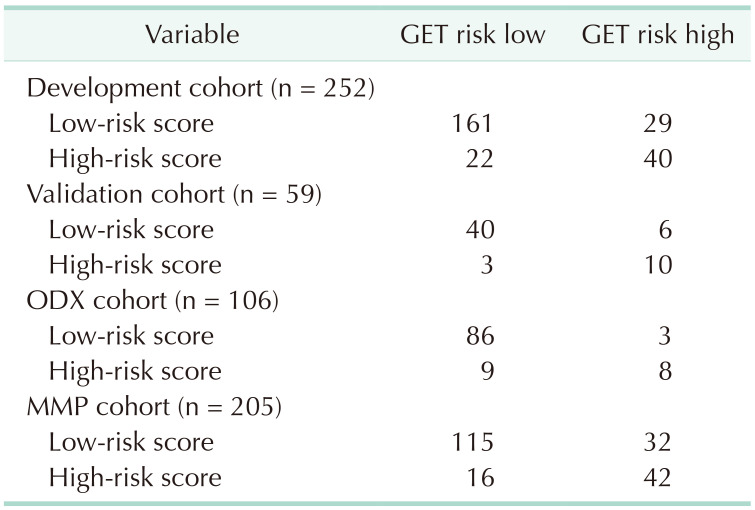
Between January 2015 and December 2022, 311 patients satisfied the inclusion criteria. A simplified risk scoring system was developed based on 75% of the patient cohort, which accounted for 252 patients (76.1%) up until June 2021. The remaining patients were used for validation. No differences exist in the clinicopathological factors between the development and validation cohorts. For simplicity in clinical practice, multivariate factors were converted into binary variables based on medical evidence or distribution. Univariate regression analysis was conducted on the development cohort, and significant associations were observed between the outcome variable and several factors, including BMI, histology, T stage, TNM stage, HG, NG, PR expression, HER2 score, and Ki-67. Multicollinearity was examined between NG and HG and between the T and TNM stages, utilizing the variance inflation factor value. A multivariate regression analysis was conducted after excluding the NG and T stages. The results of the multivariate regression analysis indicated that T stage, HG, PR score, HER2 score, and Ki-67 were significant (Table 2). The Hosmer-Lemeshow test yielded a chi-square (χ2) value of 2.686 with a P-value of 0.388. To assess the overall impact and for ease of calculation, scores were assigned to these 5 variables based on their β-coefficients. A simple scoring system was created by assigning integer values to each variable, rounded up considering their β-coefficients, and using 1 point as a reference point for a PR score of 0.613. Therefore, a score of 3 was assigned for HG 3, a score of 2 for pathologic T2 or above, and a score of 1 for a lower PR (1–20 or Alled score 3–6), HER2 2 (+), and high Ki-67 (>20) (Table 3). The sum of each score becomes the total cumulative score, representing the probability of attaining GET high risk.
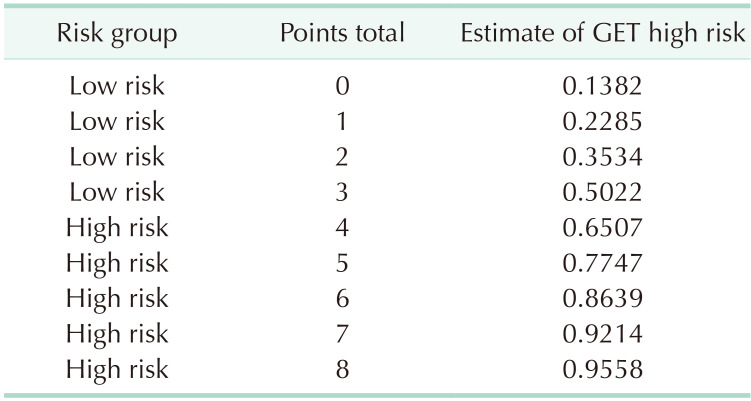
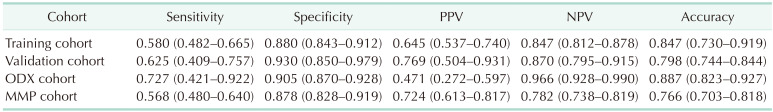
To identify the optimal cutoff point for the AUC, we analyzed Youden’s J statistic, which reached its peak at 3.6. To ascertain the estimate of GET high risk, we utilized the logistic regression formula: 1 / [1 + EXP {−(−1.83 + 0.613 × point)}], to calculate the estimate of GET high risk for each point (Table 4). When 3.6 is used as the cutoff to separate into low and high groups, there is a 65.1% probability that point 4 will be classified as high through gene expression assays (Table 5). In the training cohort, the sensitivity was 0.580 (95% confidence interval [CI], 0.482–0.665) (Table 6), specificity was 0.880 (95% CI, 0.843–0.912), positive predictive value (PPV) was 0.645 (95% CI, 0.537–0.740), negative predictive value (NPV) was 0.847 (95% CI, 0.812–0.878), and overall accuracy was 0.847 (95% CI, 0.730–0.919). In the validation cohort, the sensitivity was 0.625 (95% CI, 0.409–0.757), specificity was 0.930 (95% CI, 0.850–0.979), PPV was 0.769 (95% CI, 0.504–0.931), NPV was 0.870 (95% CI, 0.795–0.915), and overall accuracy was 0.847 (95% CI, 0.730–0.919). The receiver operating characteristic curve’s AUC value for the training set was 0.837 (95% CI, 0.710–0.964), with a P-value of less than 0.001, indicating that the AUC value was statistically significant (Fig. 1). Similarly, for the validation set, the AUC value was 0.743 (95% CI, 0.647–0.811), and it had a P-value of less than 0.001, demonstrating statistical significance (Fig. 1).
ALN metastasis plays a crucial role in patients with breast cancer, and it is a supplementary indicator for administering adjuvant systemic CTx following surgery and a prognostic factor for poor survival rates [2]. In patients with HR (+), HER2 (−) breast cancer, even when ALN metastasis is observed, a good prognosis can be expected with surgery alone, rendering adjuvant systemic CTx unnecessary. The key focus here is on the selection of patients who are suitable for these treatment strategies. Accurate patient classification enables personalized therapies while maintaining favorable oncologic outcomes. Substantial evidence from randomized controlled trials has been discussed thus far. The focus of these studies was on ODX and MMP, and they have enabled the prediction of patient prognosis based on the results of the gene signature [78].
Genomic signatures have been incorporated into the decision-making tools within the guidelines of major medical and oncology societies, such as the American Society of Clinical Oncology, the National Comprehensive Cancer Network, the St. Gallen Consensus Conference, and the European Society for Medical Oncology. However, it is challenging to administer these tests across the board. Financial burdens and issues include delays in adjuvant systemic treatment while waiting for the test results. In cases where patients have limitations with these tests, the decision to proceed with CTx is determined by factors such as anatomical stage, breast cancer subtype, Eastern Cooperative Oncology Group status, patient age, and the preferences of the patients or physicians. Even in patient groups with HR (+), HER2 (−) breast cancer, which is predicted to have a good prognosis, those with ALN metastasis tend to be classified as clinically high risk and are often considered for CTx. Due to economic concerns, disparities in medical services and unnecessary treatments should be avoided [14]. For these reasons, numerous studies have been conducted to predict genetic test results based on patient factors and traditional clinicopathological factors [15161718].
Most previous studies primarily focused on ODX or MMP. Moreover, they were conducted on all patients who underwent these tests. While a few studies measured the performance regarding the major outcome of distant recurrence in the patient population, our research focused on the results of the 2 genetic tests. Therefore, an analysis of the RxPONDER and MINDACT trial results is required to apply our research in a clinical setting. Through the RxPONDER trial, ODX has been recognized as a tool to prognosticate oncologic outcomes and predict CTx responses in patients with ALN metastases. In the case of the RxPONDER trial, all patients had ALN metastases. However, patients aged under 50 years constituted 21.5% of the total. The research results indicated that CTx could be omitted in cases with an RS of less than 25. However, caution was needed in the subgroup analysis for patients aged under 50 years. MMP is recognized as a tool for estimating survival rates after surgery. Of the 1,550 patients in the MINDACT trial, 47.6% had ALN metastases. According to the recent update on the long-term results of MINDACT, in patients with clinically high risk/genetically low risk, there is an absolute benefit of 5% ± 2.8% in the distant metastasis survival rate with additional CTx. Due to these results, there is a need to be more cautious regarding omitting CTx in patients.
We conducted a study targeting patients with breast cancer with ALN metastasis who underwent ODX or MMP. The objective was to investigate the correlation between the results of GET and traditional clinical and pathological factors. In this study, the multigene analysis revealed a strong association between high risk and the following factors: pathologic T2, HG 3, PR score (<20 or Allred score 3–6), HER2 2 (+), and Ki-67 of >20. Furthermore, a simple and efficient scoring system was developed using the estimation of β-coefficients values for each variable. This scoring system allows for easy and rapid assessment of individuals based on the identified factors, enabling a practical approach to risk stratification in clinical settings. In our study, all subjects had lymph node metastases; approximately 47% were aged under 50 years, and approximately 65% used MMP for the GET. This study only confirms the test results without survival data, such as distant metastases.
According to previous studies predicting MMP results, there was a negative correlation between MMP results and age. Our study included only patients with ALN metastases among those who underwent MMP. Moreover, it included patients who had undergone ODX, and there was no correlation with age. However, as previously described, caution is needed in patients aged under 50 years, as with the results of the RxPONDER trial. In actual clinical practice, the extent of ALN metastasis acts as a powerful factor in predicting prognosis. Macrometastases have worse outcomes than micrometastases, and the need for adjuvant CTx increases as the number of lymph node metastases increases. However, in our study, the number of metastases, and micrometastasis or macrometastasis did not correlate with the test results. According to this study, in breast cancer patients with HR (+), HER2 (−), and ALN (+), the presence or absence of micrometastasis and the number of lymph node metastases do not affect the results of the GET.
In our study, the proportion of patients with a low ER status was tiny, at just 1%. Therefore, verifying any correlation between the ER score and the results of GET was difficult. As Korean health insurance does not cover the GET and is expensive, careful consideration must be given when selecting patients for testing. Patients with low ER are likely to have a poor response to hormone therapy. Moreover, they have been diagnosed with ALN metastases. Due to the physician’s preference, they may have undergone neoadjuvant CTx or directly received adjuvant CTx without testing. As such, it is thought that these patients were likely not included in our study. Physicians at the 2 institutions conducting the research believe that if ER is low, most patients may need CTx. Therefore, there are not many patients with low ER in our study. Even excluding ER, the degree of PR expression was a significant figure in this study that included both GET. In previous studies examining ODX, PR has been reported as a strong factor that can explain up to 23% of the variability in total ODX scores. Additionally, in studies related to PR and breast cancer prognosis, the degree of PR expression has been reported to be associated with the prognosis of breast cancer [19].
Our study has several limitations. First, as it was a retrospective study, selection bias may exist. As a study design limitation, our cohort has a low proportion of low ER subjects. Moreover, the rate at which each test was used was influenced by the timing of the release of the RxPONDER and MINDACT trials. In other words, the rate of test implementation varies slightly by year. Furthermore, conducting research solely based on test results without including actual patient survival data requires caution in interpreting and applying the results in a clinical setting. Caution is necessary when applying and interpreting our study’s results for women aged under 50 years based on the findings of the RxPONDER trial. Similarly, careful consideration is essential when deciding to omit anticancer treatment based on the results of the MINDACT trial. Our study is meaningful because it includes the 2 most widely used tests, ODX and MMP, and focuses on patients with ALN metastases. Additionally, we utilized easily assessable traditional clinical pathological factors in breast cancer patients and assigned corresponding scores without complex tools. These scores can be calculated easily and quickly without sophisticated instruments. Furthermore, according to performance validation, our study showed a high NPV. Thus, our study demonstrates that it can be an excellent tool for effectively identifying individuals with a high likelihood of being predicted as negative by the GET, even if they have ALN metastases.
We developed a simple scoring system to predict GET results in patients with HR (+), HER2 (−), and ALN metastases. According to our study, a score of 3 was assigned for HG 3. Scores of 2 were given for pathologic T2 or above, while scores of 1 were assigned for PR (1–20 or Alled score 3–6), HER2 2 (+), and high Ki-67 (>20). The sum of each score becomes the total cumulative score, representing the probability of attaining GET high risk. If the total cumulative score is 4 points, there is a 65% probability that the test result will be classified as high risk. In cases with a lower score, omitting the costly decision-making test may be possible.
Notes
References
1. Chen C, Dhanda R, Tseng WY, Forsyth M, Patt DA. Evaluating use characteristics for the oncotype dx 21-gene recurrence score and concordance with chemotherapy use in early-stage breast cancer. J Oncol Pract. 2013; 9:182–187. PMID: 23942918.

2. Fisher B, Bauer M, Wickerham DL, Redmond CK, Fisher ER, Cruz AB, et al. Relation of number of positive axillary nodes to the prognosis of patients with primary breast cancer: an NSABP update. Cancer. 1983; 52:1551–1557. PMID: 6352003.

3. Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991; 19:403–410. PMID: 1757079.

4. Colditz GA, Rosner BA, Chen WY, Holmes MD, Hankinson SE. Risk factors for breast cancer according to estrogen and progesterone receptor status. J Natl Cancer Inst. 2004; 96:218–228. PMID: 14759989.

5. Choi H, Ahn SG, Bae SJ, Kim JH, Eun NL, Lee Y, et al. Comparison of programmed cell death ligand 1 status between core needle biopsy and surgical specimens of triple-negative breast cancer. Yonsei Med J. 2023; 64:518–525. PMID: 37488704.

6. Lawn AM, Frampton AE, Krell J, Waheed S, Stacey-Clear A. Lymph node ratio can further stratify prognosis in subpopulations of breast cancer patients with axillary nodal metastases. Future Oncol. 2013; 9:1425–1431. PMID: 24106893.

7. Cardoso F, van’t Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, et al. 70-Gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med. 2016; 375:717–729. PMID: 27557300.

8. Kalinsky K, Barlow WE, Gralow JR, Meric-Bernstam F, Albain KS, Hayes DF, et al. 21-Gene assay to inform chemotherapy benefit in node-positive breast cancer. N Engl J Med. 2021; 385:2336–2347. PMID: 34914339.

9. Caparica R, Brandão M, Piccart M. Systemic treatment of patients with early breast cancer: recent updates and state of the art. Breast. 2019; 48 Suppl 1:S7–S20. PMID: 31839166.

10. National Comprehensive Cancer Network (NCCN). NCCN Guidelines version 4.2023: Breast Cancer [Internet]. NCCN;2023. cited 2023 Jul 1. Available from: https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf.
11. Dowsett M, Cuzick J, Wale C, Forbes J, Mallon EA, Salter J, et al. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: a TransATAC study. J Clin Oncol. 2010; 28:1829–1834. PMID: 20212256.

12. Habel LA, Shak S, Jacobs MK, Capra A, Alexander C, Pho M, et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res. 2006; 8:R25. PMID: 16737553.

13. Andre F, Ismaila N, Allison KH, Barlow WE, Collyar DE, Damodaran S, et al. Biomarkers for adjuvant endocrine and chemotherapy in early-stage breast cancer: ASCO guideline update. J Clin Oncol. 2022; 40:1816–1837. PMID: 35439025.

14. Lim H, Kim SI, Hyun S, Lee GB, Seol A, Lee M. Uptake rate of risk-reducing salpingo-oophorectomy and surgical outcomes of female germline BRCA1/2 mutation carriers: a retrospective cohort study. Yonsei Med J. 2021; 62:1090–1097. PMID: 34816639.

15. Eaton AA, Pesce CE, Murphy JO, Stempel MM, Patil SM, Brogi E, et al. Estimating the OncotypeDX score: validation of an inexpensive estimation tool. Breast Cancer Res Treat. 2017; 161:435–441. PMID: 27928699.

16. Lee SB, Kim J, Sohn G, Kim J, Chung IY, Kim HJ, et al. A nomogram for predicting the oncotype DX recurrence score in women with T1-3N0-1miM0 hormone receptor positive, human epidermal growth factor 2 (HER2) negative breast cancer. Cancer Res Treat. 2019; 51:1073–1085. PMID: 30384581.

17. Lee YJ, Hwang YS, Kim J, Ahn SH, Son BH, Kim HJ, et al. A nomogram for predicting probability of low risk of MammaPrint results in women with clinically high-risk breast cancer. Sci Rep. 2021; 11:23509. PMID: 34873249.

18. Kim MC, Kwon SY, Choi JE, Kang SH, Bae YK. Prediction of oncotype DX recurrence score using clinicopathological variables in estrogen receptor-positive/human epidermal growth factor receptor 2-negative breast cancer. J Breast Cancer. 2023; 26:105–116. PMID: 37095618.

19. Kwak Y, Jang SY, Choi JY, Lee H, Shin DS, Park YH, et al. Progesterone receptor expression level predicts prognosis of estrogen receptor-positive/HER2-negative young breast cancer: a single-center prospective cohort study. Cancers (Basel). 2023; 15:3435. PMID: 37444546.

Fig. 1
Receiver operating characteristic curve of a simplified risk scoring system. (A) Development cohort. (B) Validation cohort. (C) Oncotype DX (Genomic Health) cohort. (D) MammaPrint (Agendia) cohort. AUC, area under the curve; CI, confidence interval.
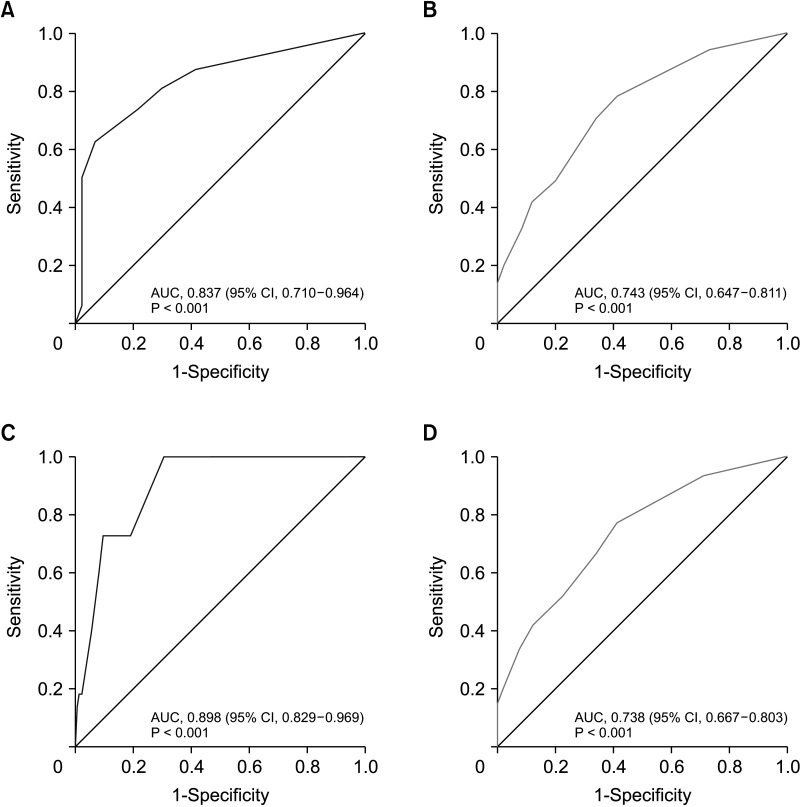
Table 2
Univariate and multivariate analyses of the predictive variables of gene expression test high risk on the development cohort




 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download