Abstract
The clinical characteristics and prognoses of acromegaly vary among patients. Assessment of current and novel predictors can lead to multilevel categorization of patients, allowing integration into new clinical guidelines and a reduction in the increased morbidity and mortality associated with acromegaly. Despite advances in the diagnosis and treatment of acromegaly, its pathophysiology remains unclear. Recent advancements in multiomics technologies, including genomics, transcriptomics, proteomics, metabolomics, and radiomics, have offered new opportunities to unravel the complex pathophysiology of acromegaly. This review comprehensively explores the emerging role of multiomics approaches in elucidating the molecular landscape of acromegaly. We discuss the potential implications of multiomics data integration in the development of novel diagnostic tools, identification of therapeutic targets, and the prospects of precision medicine in acromegaly management. By integrating diverse omics datasets, these approaches can provide valuable insights into disease mechanisms, facilitate the identification of diagnostic biomarkers, and identify potential therapeutic targets for precision medicine in the management of acromegaly.
The Songwon Medical Scientist Award is the highest scientific award of the Korean Endocrine Society to honor an individual below 45 years old who has made excellent scientific contributions and planned outstanding research proposal to progress in the field of endocrinology and metabolism. The Songwon Medical Scientist Award is named after the pen name of Professor Kab Bum Huh, who had been emphasizing the importance of research and educations in field of not only endocrinology but also overall medicine. Professor Cheol Ryong Ku received the 1st Songwon Medical Scientist Award at the 10th Seoul International Congress of Endocrinology and Metabolism of the Korean Endocrine Society in October 2022.
Approximately 15% of all primary intracranial tumors are pituitary neuroendocrine tumors [1]. Growth hormone (GH)-producing pituitary adenomas are the second most common hormone-producing pituitary tumors after prolactin-producing adenomas [1]. Acromegaly presents with various symptoms such as enlargement of the extremities and tongue, cardiac hypertrophy, osteoarthropathy, metabolic disorders, and malignancy due to prolonged excess GH [2,3]. If GH overproduction is not controlled, these complications can lead to high mortality rate [4,5].
Improvements in surgical procedures, the development of new radiotherapy techniques, and the availability of new medical therapies have resulted in considerable changes to the treatment and management of acromegaly [6,7]. However, a significant number of patients may not achieve remission owing to multiple clinical parameters [8-12]. Therefore, investigating the underlying biological mechanisms can help to identify targets for more effective treatments. This review outlines the need for a deeper understanding of patients with acromegaly to improve disease control and compliance.
Rather than examining individual genes, proteins, or metabolites, the focus shifts to analyzing complete sets: the genome, proteome, metabolome, and radiomics. These comprehensive approaches are categorized as “omics,” reflecting encompassing nature denoted by the suffix “-ome” [13]. Omics techniques are used in translational medicine, where research outcomes are translated into practical clinical applications, leveraging their inherent capacity for personalized medicine [14]. The impartial application of omics enables the evaluation of diagnosis, prognosis, and prediction of treatment responses, facilitating the customization of medical approaches, ranging from risk assessment to the handling of treatment resistance [15].
Despite recent efforts to elucidate the mechanisms and etiology of GH-secreting pituitary adenomas, the connection between genetic alterations and clinical characteristics remains unclear, and the factors influencing treatment responsiveness remain unknown. Moreover, genetic investigations specifically targeting GH-secreting pituitary tumors are limited. Therefore, a deeper understanding of the molecular basis of acromegaly is essential for developing targeted therapies and personalized treatment strategies.
In this review, we summarized the molecular aspects revealed by multiomics approaches that have shed new insight into the understanding of clinical behavior and novel therapeutic targets for patients with acromegaly and conclude with a discussion of future prospects.
Among the various omics technologies, genomics, the most established domain, involves the investigation of linked genetic variations to discover novel therapeutic reactions or disease prognoses [16]. In this context, the impact of understanding genetic variations, as observed in genome-wide association studies (GWAS), coupled with omics discoveries, has been identified [17]. Genomic studies have identified several genetic alterations associated with acromegaly, including germline variants, somatic mutations, and copy number variations.
Germline mutations occur in several known genes, including those in multiple endocrine neoplasia type 1 (MEN1), protein kinase A regulatory subunit 1α (PRKARIA), aryl hydrocarbon receptor-interacting protein (AIP), G-protein coupled receptor 101 (GPR101), cyclin dependent kinase inhibitor 1B (CDKN1B), succinate dehydrogenase subunit (SDHx), MYC associated factor X (MAX) genes, as well as familial cases with currently unknown genes [18-21].
The most commonly identified somatic mutations in acromegaly are activating mutations in the guanine nucleotide-binding protein alpha-stimulating (GNAS) gene [22]. These mutations cause a gain-of-function effect in GNAS, leading to increased production of cyclic adenosine monophosphate (cAMP), which in turn promotes cell proliferation and GH release. Notably, specific mutations have been identified at critical positions, such as codon 201, where arginine can be substituted with cysteine, histidine, or serine, and codon 227, where glutamine can be replaced with arginine or leucine [23]. Approximately 30% to 40% of sporadic acromegaly cases exhibit GNAS mutations [22]. GNAS mutations in GH-secreting pituitary tumors are associated with higher preoperative insulin-like growth factor-1 (IGF-1) levels, higher surgical remission rates, favorable responses to somatostatin analogues, and lower immediate postoperative nadir GH levels [24].
Genomic stability of GH-secreting adenomas has been extensively studied. In a GWAS involving 128 sporadic pituitary adenomas, three susceptibility loci were identified, two on chromosome 10 and one on chromosome 13 [25]. In addition, copy number variations have also been reported. Interestingly, GH-secreting pitutiary tumors do not exhibit classical oncogene mutations when analyzed by whole-genome sequencing and somatic copy number analysis, indicating potential defects in cell cycle regulation or signaling pathways [26]. Furthermore, genomic profiling of 39 GH-secreting tumors revealed heterogeneity in copy number alterations, indicating genomic instability. Some tumors exhibit substantial genomic disruption and aneuploidy, affecting up to 45% of the affected genome [27]. These tumors also harbored GNAS1 mutations. In a multiplex next-generation sequencing analysis of 16 GH-secreting pituitary tumors, copy number variations were more prevalent in GH-secreting pituitary tumors than in non-hormone-secreting pitutiary tumors [28].
Given the centrality of gene expression changes and the limitations of genetic abnormalities in GH-secreting pituitary tumors, epigenetic modifications have received considerable attention. Numerous genes involved in cell growth and signaling show altered methylation status, including cell cycle regulators such as cyclin dependent kinase inhibitor 2A (CDKN2A), CDKN1B, RB transcriptional corepressor 1 (Rb), and fibroblast growth factor receptor 2 (FGFR2) [29-31]. GH-secreting pituitary tumors display relative hypomethylation of genes involved in ion channel signaling, including voltage-gated potassium channel subunit beta-2 (KCNAB2), calcium-activated potassium channel subunit beta-4 (KCNMB4), and calcium voltage-gated channel subunit alpha1C (CACNA1C), which can be hypermethylated in nonfunctional tumors [32].
Increased global acetylation leads to increased expression of pituitary tumor-transforming gene 1 (PTTG1), bone morphogenic protein 4 (BMP4), and dopamine receptor 2 (DR2) in pituitary tumor cells. This suggests that widespread changes in epigenetic modifications could induce shifts in gene expression [33,34]. Overexpression of histone deacetylases (HDAC) within the sirtuin family (SIRT) have been observed in somatotropinomas and is correlated with smaller tumor size [35]. Peptidylarginine deaminase (PAD) enzymes plays a role in histone citrullination, a process that influences chromatin activity. The enhanced presence of PAD enzymes in somatoprolactinomas has been linked to increased mRNA targeting of the oncogenes high-mobility group A 1 (HMGA1), IGF-1, and the neuroblastoma MYC oncogene (N-MYC) by micro-RNAs (miRNAs) [36].
Epigenomic profiling can contribute to the understanding of tumorigenesis in pituitary adenomas. However, there are no significant differences in tumor subtype or prognosis.
Approximately 75% of the transcribed genes yield RNA molecules that do not participate in protein synthesis; instead, they generate noncoding RNAs [37]. These noncoding RNAs encompass miRNAs, long noncoding RNAs, circular RNAs, and P-element Induced WImpy testis in Drosophila (PIWI)-interacting RNAs [38]. Although their biological significance is not fully understood in most cases, several miRNAs have been shown to be involved in the control of pituitary tumorigenesis, aggressive features, and drug responsiveness.
miR-15a and miR-16-1 demonstrate decreased expression in GH-secreting pituitary tumors, and miR-16 is known to target GH receptor, IGF-1, IGF1 receptor, and IGF2 receptor expression [39,40]. Downregulated miRNAs were found to regulate the expression of three genes linked to proliferation: HMGA1, controlled by miR-34b and miR-548c-3p, HMGA2 regulated by miR-34b, miR-326, miR-432, miR-548c-3p, and miR-570; and E2 promoter binding factor 1 (E2F1), which is affected by miR-326 and miR-603 [41]. Overexpression of these three genes is associated with cell proliferation [41].
The dysregulation of miR-126, miR-381, and miR-338-3p controls cell proliferation, invasion, and response to therapy by affecting PTTG1 [42-44]. MiR-26b and miR-128 act on the phosphatase and tensin homologue (PTEN)/phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) pathway, which stimulates epithelial-mesenchymal transition (EMT) [45,46]. MiR-503, known as a tumor suppressor, was found to be downregulated, whereas miR-525-5p, a protector against EMT, was upregulated in GH-secreting adenomas. Dysregulation of both has been reported to control EMT during tumorigenesis [47,48].
In acromegaly, as observed in other tumors, miRNAs also play a role in resistance to treatment. For instance, miR-34a was shown to correlate with AIP expression, which is an important marker of response to somatostatin analogue treatment [49,50]. Somatostatin receptor (SSTR) 2 is regulated by miR-185 in vitro and is upregulated in non-responders to somatostatin analogues [51]. However, no direct correlation was observed between mir185 and SST2 expression in vivo. miR-125a-5p and miR-524-5p are decreased in response to somatostatin analogues [42]. miR-866-5p, a type of oncomiR, is upregulated In responsive tumors [42].
Transcriptomics, an additional high-throughput methodology, involves concurrent qualitative and quantitative analyses of specific mRNA species and focuses on dysregulated genes in acromegaly [52]. The integration of transcriptomic data provides a comprehensive understanding of tumor aggressiveness and treatment responsiveness in patients with acromegaly.
Signal transducer and activator of transcription 3 (STAT3) is a signaling molecule that has been implicated in GH hypersecretion by somatotrophic cells and therefore holds potential as a therapeutic target for GH-secreting pituitary tumors. GH-secreting tumors exhibit increased STAT3 expression, which leads to enhanced GH synthesis [53]. Blocking STAT3 activity inhibits the growth of somatotroph tumor xenografts and suppresses GH secretion in cultured cells derived from human GH-secreting pituitary tumors [53].
PTTG regulates cell cycle by interacting with p53 [54]. More than 70% of GH-secreting pituitary tumors exhibit elevated PTTG expression, which significantly contributes to cellular senescence [55]. PTTG expression may be indicative of aggressiveness in various types of pituitary adenoma [56].
Microarray analysis of GH-secreting pituitary tumors identified E-cadherin mRNA expression to be negatively associated with tumor size and invasiveness, and positively associated with GH and IGF-I levels in serum and response to somatostatin analogues [57]. Several genes, including epithelial splicing regulatory protein 1 (ESRP1), pleomorphic adenoma gene 1 (PLAG1) like zinc finger 1 (ZAC1), and beta arrestin-1, is reported as predictors of responsiveness to medical treatment [57-60].
Rapid polymerase chain reaction amplification of cDNA ends using human pituitary tumors identified a novel alternatively spliced somatostatin 5 (sst5) receptor variant [61]. The presence of truncated sst5 (sst5TMD4) is associated with increased aggressive features and worse prognosis in GH-secreting pituitary tumors [62]. Enhanced p21 expression is linked to less aggressive GH-secreting pituitary tumors and likely exerts a mitigating effect on cell proliferation [21].
Some studies have identified somatic genetic alterations in GH-secreting pituitary tumors using whole-exome sequencing. Whole-exome sequencing detected arm-level somatic copy number alterations (SCNA) in 42 pituitary macroadenoma samples at extensive sites across the genome in 29% of the specimens. Chromosomal alterations are more frequent in GH-secreting pituitary tumors [63,64]. Both cAMP and Fanconi anaemia DNA damage repair pathways are affected by SCNA in GH-secreting pituitary tumors [63]. Another study reported that in addition to the cAMP pathway, calcium signaling might be involved in the pathogenesis of GH-secreting pituitary tumors [64]. One study proposed novel somatic variants (48 genes with 59 variants) in patients with acromegaly with neither GNAS variants nor family history [65]. Single cell RNA sequencing of pituitary-specific positive transcription factor 1 positive pituitary adenomas (PIT1-PAs) proposed interferon-γ and cadherin 2-targeted drugs are promising therapeutic methods for aggressive PIT1-PAs [66].
Proteomics is another omics-related technology that examines the protein content of an organism, tissue, or cell to understand the function or structure of a specific protein. This technology can be used in various research settings with different capacities to identify diagnostic markers, produce vaccines, and even interpret the protein pathways of disorders [67]. Herein, large-scale protein characterisation using this high-throughput proteomic strategy may benefit from mass spectroscopy techniques [68].
Although limited, proteomics studies have been conducted in patients with acromegaly. ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 (ATP2A2) and AT-rich interaction domain 5 B (ARID5B) correlated with the GH change rate in the octreotide loading test, and WWC family member 3 (WWC3), serine incorporator 1 (SERINC1), and zinc finger AN1-type containing 3 (ZFAND3) correlated with the tumor volume change rate after somatostatin analogue treatment [69]. One study evaluated biomarkersare changed after surgical treatment and revealed intensities of two isoforms of transthyretin, haptoglobin (HP) a2, haemoglobin β subunit (HBB) and two isoforms of apolipoprotein A1 (APOA1), and complement C4B (C4B) precursor could be used as potential biomarkers [70].
Metabolomics is a rapidly growing technology that has identified perturbations in various metabolic pathways, including glucose, lipid, and amino acid metabolism. These findings may contribute to the development of novel diagnostic biomarkers and targeted therapies for acromegaly.
Feng et al. [7] reported that all pituitary adenomas, including GH-secreting pituitary adenomas, display downregulated glucose metabolism and glycolysis compared to normal tissues. They revealed that isocitrate dehydrogenase (NADP(+)) 2 (IDH2) is a key player in the reprogrammed metabolism of such tumors, and confirmed that IDH2 is a potential target for inhibiting tumor cell growth and secretion by blocking IDH2 [71].
Radiomics transforms radiological images into extractable data by automatically quantifying comprehensive imaging characteristics [75]. Previous studies have shown promising results in predicting the molecular state, severity, and outlook of gliomas [76]. Recent investigations have also shown encouraging findings for predicting the subtype or treatment response of pituitary adenomas. Although basic observations, such as cavernous sinus invasion on magnetic resonance imaging (MRI), are also associated with prognosis, advancements in MRI methodologies have led to a variety of ongoing radiomic studies [77,78].
Park et al. [79,80] proposed that radiomics features have the potential to predict the granulation pattern of GH-secreting pituitary tumors. Earlier studies have suggested that densely granulated adenomas respond more favorably to somatostatin analogue treatment than sparsely granulated adenomas do. This is particularly relevant, given the limited accessibility of electron microscopy in clinical settings.
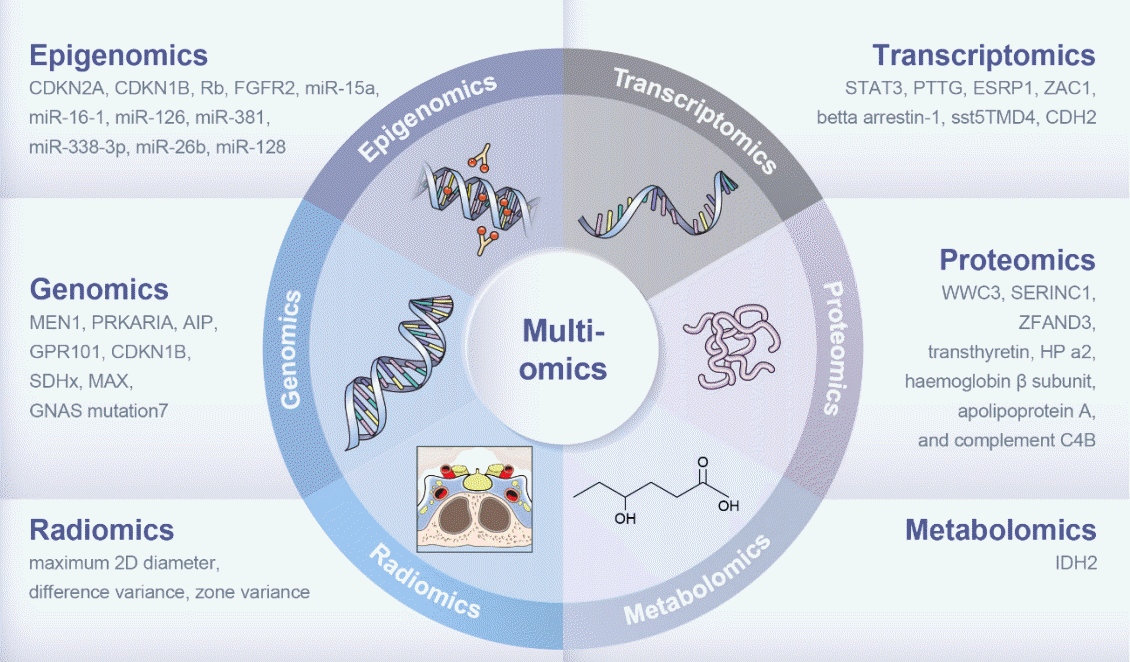
Nucleic acid-based methods, including genomics, epigenomics, and transcriptomics have been extensively used in patients with acromegaly, providing information on multiomics characteristics related to variable parameters such as drug responsiveness and recurrence (Fig. 1). To date, studies have focused on single-omics approaches to classify patients with acromegaly. Integrating various omics datasets offers a comprehensive perspective that enables us to derive more precise insights to understanding diseases and searching possible targets for treatment. Multiomics strategies have enlightened us in a manner that singular omics approaches alone cannot achieve. The integration of clinical data such as treatment response and disease outcomes through a multiomics approach can facilitate the identification of personalized treatment strategies. Further research and collaborative efforts are required to translate these findings into clinical practice.
ACKNOWLEDGMENTS
This work was supported by the Korean Endocrine Society for Songwon Award 2022. We would like to thank Professor Kab Bum Huh for encouraging and supporting young endocrinologists to start and continue the research. Medical Illustration & Design (MID), as a member of the Medical Research Support Services of Yonsei University College of Medicine, providing excellent support with medical illustration.
REFERENCES
1. Chanson P, Salenave S, Kamenicky P, Cazabat L, Young J. Pituitary tumours: acromegaly. Best Pract Res Clin Endocrinol Metab. 2009; 23:555–74.
2. Kim J, Hong N, Choi J, Moon JH, Kim EH, Hong JW, et al. Sex differences in mortality in patients with acromegaly: a nationwide cohort study in Korea. Eur J Endocrinol. 2023; 189:225–34.

3. Park KH, Lee EJ, Seo GH, Ku CR. Risk for acromegaly-related comorbidities by sex in Korean acromegaly. J Clin Endocrinol Metab. 2020; 105:dgz317.

4. Gadelha MR, Kasuki L, Lim DS, Fleseriu M. Systemic complications of acromegaly and the impact of the current treatment landscape: an update. Endocr Rev. 2019; 40:268–332.

5. Hwang YA, Lee HW, Ahn SH, Lee EJ, Ku CR, Kim SU. Positive association between nonalcoholic fatty liver disease and growth hormone deficiency in patients with nonfunctioning pituitary adenoma. Front Endocrinol (Lausanne). 2023; 13:1057769.

6. Ribeiro-Oliveira A Jr, Barkan A. The changing face of acromegaly: advances in diagnosis and treatment. Nat Rev Endocrinol. 2012; 8:605–11.
7. Langlois F, McCartney S, Fleseriu M. Recent progress in the medical therapy of pituitary tumors. Endocrinol Metab (Seoul). 2017; 32:162–70.

8. Ku CR, Kim EH, Oh MC, Lee EJ, Kim SH. Surgical and endocrinological outcomes in the treatment of growth hormonesecreting pituitary adenomas according to the shift of surgical paradigm. Neurosurgery. 2012; 71(2 Suppl Operative):ons 192–203.

9. Jung IH, Choi S, Ku CR, Lee SG, Lee EJ, Kim SH, et al. Revisiting the role of insulin-like growth factor-1 measurement after surgical treatment of acromegaly. J Clin Endocrinol Metab. 2021; 106:e2589–99.

10. Kim J, Hwang YA, Park YW, Moon JH, Kim EH, Hong JW, et al. Revisiting growth hormone nadir cut-offs for remission in patients with acromegaly. Eur J Endocrinol. 2022; 186:657–65.

11. Park SH, Ku CR, Moon JH, Kim EH, Kim SH, Lee EJ. Age- and sex-specific differences as predictors of surgical remission among patients with acromegaly. J Clin Endocrinol Metab. 2018; 103:909–16.

12. Kim EH, Oh MC, Lee EJ, Kim SH. Predicting long-term remission by measuring immediate postoperative growth hormone levels and oral glucose tolerance test in acromegaly. Neurosurgery. 2012; 70:1106–13.

13. Yan J, Risacher SL, Shen L, Saykin AJ. Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Brief Bioinform. 2018; 19:1370–81.

15. Ku CR, Melnikov V, Zhang Z, Lee EJ. Precision therapy in acromegaly caused by pituitary tumors: how close is it to reality? Endocrinol Metab (Seoul). 2020; 35:206–16.

17. Uffelmann E, Huang QQ, Munung NS, De Vries J, Okada Y, Martin AR, et al. Genome-wide association studies. Nat Rev Methods Primers. 2021; 1:59.

18. Trivellin G, Daly AF, Faucz FR, Yuan B, Rostomyan L, Larco DO, et al. Gigantism and acromegaly due to Xq26 microduplications and GPR101 mutation. N Engl J Med. 2014; 371:2363–74.

19. Gadelha MR, Kasuki L, Korbonits M. The genetic background of acromegaly. Pituitary. 2017; 20:10–21.

20. Caimari F, Korbonits M. Novel genetic causes of pituitary adenomas. Clin Cancer Res. 2016; 22:5030–42.

21. Gillam MP, Ku CR, Lee YJ, Kim J, Kim SH, Lee SJ, et al. Somatotroph-specific Aip-deficient mice display pretumorigenic alterations in cell-cycle signaling. J Endocr Soc. 2017; 1:78–95.

22. Landis CA, Masters SB, Spada A, Pace AM, Bourne HR, Vallar L. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature. 1989; 340:692–6.

23. Landis CA, Harsh G, Lyons J, Davis RL, McCormick F, Bourne HR. Clinical characteristics of acromegalic patients whose pituitary tumors contain mutant Gs protein. J Clin Endocrinol Metab. 1990; 71:1416–20.
24. Jung H, Kim K, Kim D, Moon JH, Kim EH, Kim SH, et al. Associations of GNAS mutations with surgical outcomes in patients with growth hormone-secreting pituitary adenoma. Endocrinol Metab (Seoul). 2021; 36:342–50.

25. Ye Z, Li Z, Wang Y, Mao Y, Shen M, Zhang Q, et al. Common variants at 10p12.31, 10q21.1 and 13q12.13 are associated with sporadic pituitary adenoma. Nat Genet. 2015; 47:793–7.

26. Lecoq AL, Kamenicky P, Guiochon-Mantel A, Chanson P. Genetic mutations in sporadic pituitary adenomas: what to screen for? Nat Rev Endocrinol. 2015; 11:43–54.

27. Hage M, Viengchareun S, Brunet E, Villa C, Pineau D, Bouligand J, et al. Genomic alterations and complex subclonal architecture in sporadic GH-secreting pituitary adenomas. J Clin Endocrinol Metab. 2018; 103:1929–39.

28. Bi WL, Greenwald NF, Ramkissoon SH, Abedalthagafi M, Coy SM, Ligon KL, et al. Clinical identification of oncogenic drivers and copy-number alterations in pituitary tumors. Endocrinology. 2017; 158:2284–91.

29. Yoshino A, Katayama Y, Ogino A, Watanabe T, Yachi K, Ohta T, et al. Promoter hypermethylation profile of cell cycle regulator genes in pituitary adenomas. J Neurooncol. 2007; 83:153–62.

30. Kirsch M, Morz M, Pinzer T, Schackert HK, Schackert G. Frequent loss of the CDKN2C (p18INK4c) gene product in pituitary adenomas. Genes Chromosomes Cancer. 2009; 48:143–54.
31. Hossain MG, Iwata T, Mizusawa N, Qian ZR, Shima SW, Okutsu T, et al. Expression of p18(INK4C) is down-regulated in human pituitary adenomas. Endocr Pathol. 2009; 20:114–21.

32. Ling C, Pease M, Shi L, Punj V, Shiroishi MS, Commins D, et al. A pilot genome-scale profiling of DNA methylation in sporadic pituitary macroadenomas: association with tumor invasion and histopathological subtype. PLoS One. 2014; 9:e96178.

33. Yacqub-Usman K, Duong CV, Clayton RN, Farrell WE. Preincubation of pituitary tumor cells with the epidrugs zebularine and trichostatin A are permissive for retinoic acid-augmented expression of the BMP-4 and D2R genes. Endocrinology. 2013; 154:1711–21.

34. Li T, Huang H, Huang B, Huang B, Lu J. Histone acetyltransferase p300 regulates the expression of human pituitary tumor transforming gene (hPTTG). J Genet Genomics. 2009; 36:335–42.

35. Grande IP, Amorim PV, Freire AC, Jallad RS, Musolino NR, Cescato VA, et al. Differential gene expression of sirtuins between somatotropinomas and nonfunctioning pituitary adenomas. Pituitary. 2018; 21:355–61.

36. DeVore SB, Young CH, Li G, Sundararajan A, Ramaraj T, Mudge J, et al. Histone citrullination represses microRNA expression, resulting in increased oncogene mRNAs in somatolactotrope cells. Mol Cell Biol. 2018; 38:e00084–18.

37. Zhang P, Wu W, Chen Q, Chen M. Non-coding RNAs and their integrated networks. J Integr Bioinform. 2019; 16:20190027.

38. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012; 482:339–46.

39. Bottoni A, Piccin D, Tagliati F, Luchin A, Zatelli MC, degli Uberti EC. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J Cell Physiol. 2005; 204:280–5.

40. Elzein S, Goodyer CG. Regulation of human growth hormone receptor expression by microRNAs. Mol Endocrinol. 2014; 28:1448–59.

41. D’Angelo D, Palmieri D, Mussnich P, Roche M, Wierinckx A, Raverot G, et al. Altered microRNA expression profile in human pituitary GH adenomas: down-regulation of miRNA targeting HMGA1, HMGA2, and E2F1. J Clin Endocrinol Metab. 2012; 97:E1128–38.
42. Mao ZG, He DS, Zhou J, Yao B, Xiao WW, Chen CH, et al. Differential expression of microRNAs in GH-secreting pituitary adenomas. Diagn Pathol. 2010; 5:79.

43. Lee YJ, Cho JM, Moon JH, Ku CR, Kim J, Kim SH, et al. Increased miR-338-3p expression correlates with invasiveness of GH-producing pituitary adenomas. Endocrine. 2017; 58:184–9.

44. Salehi F, Kovacs K, Scheithauer BW, Lloyd RV, Cusimano M. Pituitary tumor-transforming gene in endocrine and other neoplasms: a review and update. Endocr Relat Cancer. 2008; 15:721–43.

45. Palumbo T, Faucz FR, Azevedo M, Xekouki P, Iliopoulos D, Stratakis CA. Functional screen analysis reveals miR-26b and miR-128 as central regulators of pituitary somatomammotrophic tumor growth through activation of the PTENAKT pathway. Oncogene. 2013; 32:1651–9.

46. Jiang N, Dai Q, Su X, Fu J, Feng X, Peng J. Role of PI3K/AKT pathway in cancer: the framework of malignant behavior. Mol Biol Rep. 2020; 47:4587–629.

47. Xie P, Han Q, Liu D, Yao D, Lu X, Wang Z, et al. miR-525-5p modulates proliferation and epithelial-mesenchymal transition of glioma by targeting Stat-1. Onco Targets Ther. 2020; 13:9957–66.
48. Gupta G, Chellappan DK, de Jesus Andreoli Pinto T, Hansbro PM, Bebawy M, Dua K. Tumor suppressor role of miR-503. Panminerva Med. 2018; 60:17–24.

49. Denes J, Kasuki L, Trivellin G, Colli LM, Takiya CM, Stiles CE, et al. Regulation of aryl hydrocarbon receptor interacting protein (AIP) protein expression by MiR-34a in sporadic somatotropinomas. PLoS One. 2015; 10:e0117107.

50. Bogner EM, Daly AF, Gulde S, Karhu A, Irmler M, Beckers J, et al. miR-34a is upregulated in AIP-mutated somatotropinomas and promotes octreotide resistance. Int J Cancer. 2020; 147:3523–38.
51. Fan X, Mao Z, He D, Liao C, Jiang X, Lei N, et al. Expression of somatostatin receptor subtype 2 in growth hormone-secreting pituitary adenoma and the regulation of miR-185. J Endocrinol Invest. 2015; 38:1117–28.

52. Hegde PS, White IR, Debouck C. Interplay of transcriptomics and proteomics. Curr Opin Biotechnol. 2003; 14:647–51.

53. Zhou C, Jiao Y, Wang R, Ren SG, Wawrowsky K, Melmed S. STAT3 upregulation in pituitary somatotroph adenomas induces growth hormone hypersecretion. J Clin Invest. 2015; 125:1692–702.

54. Bernal JA, Luna R, Espina A, Lazaro I, Ramos-Morales F, Romero F, et al. Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis. Nat Genet. 2002; 32:306–11.

55. Chesnokova V, Zonis S, Kovacs K, Ben-Shlomo A, Wawrowsky K, Bannykh S, et al. p21(Cip1) restrains pituitary tumor growth. Proc Natl Acad Sci U S A. 2008; 105:17498–503.
56. Filippella M, Galland F, Kujas M, Young J, Faggiano A, Lombardi G, et al. Pituitary tumour transforming gene (PTTG) expression correlates with the proliferative activity and recurrence status of pituitary adenomas: a clinical and immunohistochemical study. Clin Endocrinol (Oxf). 2006; 65:536–43.

57. Lekva T, Berg JP, Fougner SL, Olstad OK, Ueland T, Bollerslev J. Gene expression profiling identifies ESRP1 as a potential regulator of epithelial mesenchymal transition in somatotroph adenomas from a large cohort of patients with acromegaly. J Clin Endocrinol Metab. 2012; 97:E1506–14.

58. Gatto F, Biermasz NR, Feelders RA, Kros JM, Dogan F, van der Lely AJ, et al. Low beta-arrestin expression correlates with the responsiveness to long-term somatostatin analog treatment in acromegaly. Eur J Endocrinol. 2016; 174:651–62.

59. Theodoropoulou M, Zhang J, Laupheimer S, Paez-Pereda M, Erneux C, Florio T, et al. Octreotide, a somatostatin analogue, mediates its antiproliferative action in pituitary tumor cells by altering phosphatidylinositol 3-kinase signaling and inducing Zac1 expression. Cancer Res. 2006; 66:1576–82.

60. Chahal HS, Trivellin G, Leontiou CA, Alband N, Fowkes RC, Tahir A, et al. Somatostatin analogs modulate AIP in somatotroph adenomas: the role of the ZAC1 pathway. J Clin Endocrinol Metab. 2012; 97:E1411–20.

61. Duran-Prado M, Gahete MD, Martinez-Fuentes AJ, Luque RM, Quintero A, Webb SM, et al. Identification and characterization of two novel truncated but functional isoforms of the somatostatin receptor subtype 5 differentially present in pituitary tumors. J Clin Endocrinol Metab. 2009; 94:2634–43.

62. Luque RM, Ibanez-Costa A, Neto LV, Taboada GF, Hormaechea-Agulla D, Kasuki L, et al. Truncated somatostatin receptor variant sst5TMD4 confers aggressive features (proliferation, invasion and reduced octreotide response) to somatotropinomas. Cancer Lett. 2015; 359:299–306.

63. Ben-Shlomo A, Deng N, Ding E, Yamamoto M, Mamelak A, Chesnokova V, et al. DNA damage and growth hormone hypersecretion in pituitary somatotroph adenomas. J Clin Invest. 2020; 130:5738–55.

64. Ronchi CL, Peverelli E, Herterich S, Weigand I, Mantovani G, Schwarzmayr T, et al. Landscape of somatic mutations in sporadic GH-secreting pituitary adenomas. Eur J Endocrinol. 2016; 174:363–72.

65. Ku CR, Lim H, Lee YJ, Kim SH, Kim D, Kim SH, et al. Novel somatic variants involved in biochemical activity of pure growth hormone-secreting pituitary adenoma without GNAS variant. Sci Rep. 2021; 11:16530.

66. Lyu L, Jiang Y, Ma W, Li H, Liu X, Li L, et al. Single-cell sequencing of PIT1-positive pituitary adenoma highlights the pro-tumour microenvironment mediated by IFN-γ-induced tumour-associated fibroblasts remodelling. Br J Cancer. 2023; 128:1117–33.

67. Aslam B, Basit M, Nisar MA, Khurshid M, Rasool MH. Proteomics: technologies and their applications. J Chromatogr Sci. 2017; 55:182–96.

68. Zhang Z, Wu S, Stenoien DL, Pasa-Tolic L. High-throughput proteomics. Annu Rev Anal Chem (Palo Alto Calif). 2014; 7:427–54.

69. Yamato A, Nagano H, Gao Y, Matsuda T, Hashimoto N, Nakayama A, et al. Proteogenomic landscape and clinical characterization of GH-producing pituitary adenomas/somatotroph pituitary neuroendocrine tumors. Commun Biol. 2022; 5:1304.

70. Cruz-Topete D, Christensen B, Sackmann-Sala L, Okada S, Jorgensen JO, Kopchick JJ. Serum proteome changes in acromegalic patients following transsphenoidal surgery: novel biomarkers of disease activity. Eur J Endocrinol. 2011; 164:157–67.

71. Feng J, Gao H, Zhang Q, Zhou Y, Li C, Zhao S, et al. Metabolic profiling reveals distinct metabolic alterations in different subtypes of pituitary adenomas and confers therapeutic targets. J Transl Med. 2019; 17:291.

72. Oklu R, Deipolyi AR, Wicky S, Ergul E, Deik AA, Chen JW, et al. Identification of small compound biomarkers of pituitary adenoma: a bilateral inferior petrosal sinus sampling study. J Neurointerv Surg. 2014; 6:541–6.

73. Feng J, Zhang Q, Zhou Y, Yu S, Hong L, Zhao S, et al. Integration of proteomics and metabolomics revealed metabolite-protein networks in ACTH-secreting pituitary adenoma. Front Endocrinol (Lausanne). 2018; 9:678.

74. Ijare OB, Holan C, Hebert J, Sharpe MA, Baskin DS, Pichumani K. Elevated levels of circulating betahydroxybutyrate in pituitary tumor patients may differentiate prolactinomas from other immunohistochemical subtypes. Sci Rep. 2020; 10:1334.

75. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016; 278:563–77.

76. Park YW, Choi YS, Ahn SS, Chang JH, Kim SH, Lee SK. Radiomics MRI phenotyping with machine learning to predict the grade of lower-grade gliomas: a study focused on nonenhancing tumors. Korean J Radiol. 2019; 20:1381–9.

77. Park HH, Kim EH, Ku CR, Lee EJ, Kim SH. Outcomes of aggressive surgical resection in growth hormone-secreting pituitary adenomas with cavernous sinus invasion. World Neurosurg. 2018; 117:e280–9.

78. Kim EH, Oh MC, Chang JH, Moon JH, Ku CR, Chang WS, et al. Postoperative gamma knife radiosurgery for cavernous sinus-invading growth hormone-secreting pituitary adenomas. World Neurosurg. 2018; 110:e534–45.

Fig. 1.
Presentative biomarkers revealed by multiomics approach in the patients with acromegaly. CDKN2A, cyclin dependent kinase inhibitor 2A; CDKN1B, cyclin dependent kinase inhibitor 1B; Rb, RB transcriptional corepressor 1; FGFR2, fibroblast growth factor receptor 2; miR, micro-RNA; STAT3, signal transducer and activator of transcription 3; PTTG, pituitary tumor-transforming gene; ESRP1, epithelial splicing regulatory protein 1; ZAC1, pleomorphic adenoma gene 1 like zinc finger 1; sst5TMD4, truncated spliced somatostatin 5; CDH2, cadherin 2; MEN1, multiple endocrine neoplasia type 1; PRKARIA, protein kinase A regulatory subunit 1α; AIP, aryl hydrocarbon receptorinteracting protein; GPR101, G-protein coupled receptor 101; SDHx, succinate dehydrogenase subunit; MAX, MYC associated factor X; GNAS, guanine nucleotide-binding protein alpha-stimulating; WWC3, WWC family member 3; SERINC1, serine incorporator 1; ZFAND3, zinc finger AN1-type containing 3; HP, haptoglobin; 2D, two-dimention; IDH 2, isocitrate dehydrogenase (NADP(+)) 2.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download