INTRODUCTION
MATERIALS AND METHODS
Study population
Nucleic acid extraction from plasma
Cell-free DNA NGS assays
Table 1
| Mutation type | Genes | |
|---|---|---|
| Oncomine Pan-Cancer Cell-Free Assay | AlphaLiquid 100 kit | |
| SNVs, small indels | AKT1, ALK, APC, AR, ARAF, BRAF, BRCA1*, BRCA2*, CHEK2, CTNNB1, DDR2, EGFR, ERBB2, ERBB3, ESR1, FBXW7, FGFR1, FGFR2, FGFR3, FGFR4, FLT3, GNA11, GNAQ, GNAS, HRAS, IDH1, IDH2, KIT, KRAS, MAP2K1, MAP2K2, MET, MTOR, NRAS, NTRK1, NTRK3, PDGFRA, PIK3CA, PTEN, RAF1, RET, ROS1, SF3B1, SMAD4, SMO, TP53 | ABL1, AKT1, AKT2, ALK, APC, AR, ARAF, ARID1A, ATM, BRAF, BRCA1, BRCA2, BTK, CBL, CCND1, CCND2, CCNE1, CD274, CDH1, CDK4, CDK6, CDKN2A, CEBPA, CSF1R, CTNNB1, DDR2, DPYD, EGFR, ERBB2, ERBB3, ESR1, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GATA3, GNA11, GNAQ, GNAS, HRAS, IDH1, IDH2, IGF1R, JAK2, JAK3, KDM6A, KDR, KEAP1, KIT, KRAS, MAP2K1, MAP2K2, MAPK1, MAPK3, MDM2, MET, MLH1, MPL, MSH2, MSH6, MTOR, MYC, MYCN, NF1, NF2, NFE2L2, NOTCH1, NPM1, NRAS, NTRK1, NTRK2, NTRK3, PDCD1LG2, PDGFRA, PDGFRB, PIK3CA, PIK3R1, PMS2, PPP2R1A, PTEN, PTPN11, RAF1, RB1, RET, RHEB, RHOA, RIT1, RNF43, ROS1, RUNX1, SETD2, SMAD4, SMO, STAG2, STK11, TCF7L2, TERT, TOP2A, TP53, TSC1, TSC2, U2AF1, UGT1A1, VHL |
| CNVs | CCND1, CCND2, CCND3, CDK4, CDK6, EGFR, ERBB2, FGFR1, FGFR2, FGFR3, MET, MYC, MYCN* | ABL1, AKT1, AKT2, ALK, APC, AR, ARAF, ARID1A, ATM, BRAF, BRCA1, BRCA2, BTK, CBL, CCND1, CCND2, CCNE1, CD274, CDH1, CDK4, CDK6, CDKN2A, CEBPA, CSF1R, CTNNB1, DDR2, DPYD, EGFR, ERBB2, ERBB3, ESR1, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GATA3, GNA11, GNAQ, GNAS, HRAS, IDH1, IDH2, IGF1R, JAK2, JAK3, KDM6A, KDR, KEAP1, KIT, KRAS, MAP2K1, MAP2K2, MAPK1, MAPK3, MDM2, MET, MLH1, MPL, MSH2, MSH6, MTOR, MYC, MYCN, NF1, NF2, NFE2L2, NOTCH1, NPM1, NRAS, NTRK1, NTRK2, NTRK3, PDCD1LG2, PDGFRA, PDGFRB, PIK3CA, PIK3R1, PMS2, PPP2R1A, PTEN, PTPN11, RAF1, RB1, RET, RHEB, RHOA, RIT1, RNF43, ROS1, RUNX1, SETD2, SMAD4, SMO, STAG2, STK11, TCF7L2, TERT, TOP2A, TP53, TSC1, TSC2, U2AF1, UGT1A1, VHL |
| Fusions | ALK, BRAF, ERG, ETV1, FGFR1, FGFR2, FGFR3, MET, NTRK1, NTRK3, RET, ROS1 | ALK, BCR, BRAF, EGFR, FGFR2, FGFR3, NTRK1, NTRK2, RET, ROS1 |
| Exon skipping | MET exon 14 skipping | MET exon 14 skipping |
Assay A
Assay B
Data interpretation and reporting
RESULTS
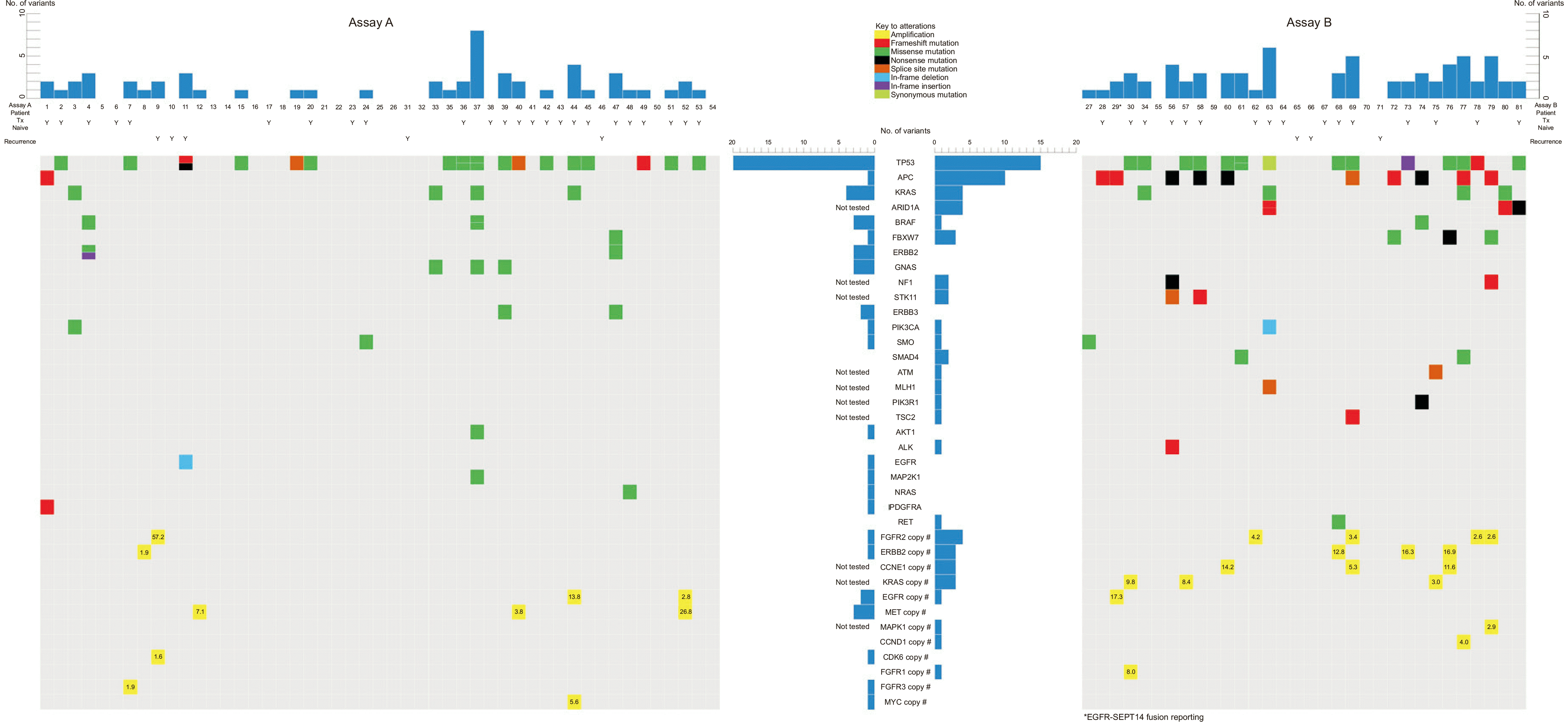
Quality metrics
Genomic alterations in GC
Table 2
| Gene | Frequency | N by mutation type |
|---|---|---|
| TP53 | 38.3% (31/81) | 31 SNVs and 4 indels |
| APC | 13.6% (11/81) | 5 SNVs and 6 indels |
| KRAS | 9.9% (8/81) | 8 SNVs |
| ARID1A | 9.4% (3/32) | 1 SNV and 3 indels |
| FBXW7 | 4.9% (4/81) | 4 SNVs |
| BRAF | 3.7% (3/81) | 4 SNVs |
| FGFR2* | 6.2% (5/81) | 5 amplifications |
| ERBB2* | 4.9% (4/81) | 4 amplifications |
| CCNE1* | 9.4% (3/32) | 3 amplifications |
| KRAS* | 9.4% (3/32) | 3 amplifications |
| EGFR* | 3.7% (3/81) | 3 amplifications |
| MET* | 3.7% (3/81) | 3 amplifications |
Molecular characterization for targeted therapy in clinical trials
Table 3
| Biomarker* | Established/in clinical trials‡ | N | Specifications |
|---|---|---|---|
| ERBB2 amplification |
First line: trastuzumab+chemotherapy Third line: trastuzumab, deruxtecan NCT05190445, NCT05152147, NCT02465060, NCT04143711 |
4 (4/81, 4.9%) |
Assay A: 1/49, 2.0% Assay B: 3/32, 9.4% |
| FGFR2 amplification | NCT05019794, NCT04189445 | 5 (5/81, 6.2%) |
Assay A: 1/49, 2.0% Assay B: 4/32, 12.5% |
| FGFR1 amplification | NCT05019794, NCT04189445 | 1 (1/81, 1.2%) |
Assay A: 0/49, 0.0% Assay B: 1/32, 3.1% |
| EGFR amplification | NCT04077255, NCT04739202 | 3 (3/81, 3.7%) |
Assay A: 2/49, 4.1% Assay B: 1/32, 3.1% |
| CCNE1 amplification† | NCT05252416 | 3 (3/32, 9.4%) | - |
| RAS mutation or amplification† | NCT02465060 | 12 (9/81, 11.1%; 3/32, 9.4%) |
Assay A: 5/49, 10.2% Assay B: 4/32, 12.5% 9 mutations (8 KRAS, 1 NRAS) 3 KRAS amplifications |
| TP53 mutation | NCT03641313 | 31 (31/81, 38.3%) |
Assay A: 17/49, 34.7% Assay B: 14/32, 43.8% 35 mutations in 31 patients |
| PIK3CA mutation or amplification† | NCT04526470, NCT04739202, NCT02465060 | 2 (2/81, 2.5%) |
Assay A: 1/49, 2.0% Assay B: 1/32, 3.1% 2 mutations No amplification |
| ARID1A mutation† | NCT05379972 | 3 (3/32, 9.4%) | 4 mutations from 3 patients |
| MET amplification | NCT04923932, NCT05620628, NCT05439993, NCT03993873, NCT02465060 | 3 (3/81, 3.7%) |
Assay A: 3/49, 6.1% Assay B: 0/32, 0.0% |
| TSC2 null† | NCT02465060 | 1 (1/32, 3.1%) | p.Phe471LeufsTer14 from #69 patient |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download