Abstract
Background
Staphylococcal cassette chromosome mec type V (SCCmec V) methicillin-resistant Staphylococcus aureus (MRSA) has been recovered from patients and livestock. Using comparative genomic analyses, we evaluated the phylogenetic emergence of SCCmec V after transmission from overseas donor strains to Korean recipient strains.
Methods
Sixty-three complete MRSA SCCmec V genomes (including six Korean clinical isolates) were used to construct a phylogenetic tree. Single-nucleotide polymorphisms were identified using Snippy, and a maximum-likelihood-based phylogenetic tree was constructed using RAxML. The possible emergence of the most common ancestor was estimated using BactDating. To estimate mecA horizontal gene transfer (HGT) events, Ranger-dtl was applied to 818 SCCmec V strains using publicly available whole-genome data.
Results
The phylogenetic tree showed five major clades. German strains formed a major clade; their possible origin was traced to the 1980s. The emergence of Korean SCCmec V clinical isolates was traced to 2000–2010. mecA HGT events in Staphylococcus spp. were identified in seven strains. P7 (Hong Kong outbreak strain) served as the donor strain for two Korean sequence type (ST) 59 strains, whereas the other five recipient strains emerged from different SCCmec V donors.
Conclusions
Most Korean SCCmec V strains may have emerged during 2000–2010. A unique MRSA SCCmec V strain, ST72 (a Korean common type of community-associated MRSA), was also identified. The genomic dynamics of this clone with a zoonotic background should be monitored to accurately understand MRSA evolution.
Methicillin-resistant Staphylococcus aureus (MRSA) is among the most prevalent nosocomial pathogens in Korea, where it was first discovered in 1969 [1]. MRSA accounted for 25% of S. aureus isolates obtained from Korean tertiary care hospitals in 1980, 54% in 1990, and 70% in 2000 [2, 3]. Korean invasive or non-invasive community-acquired MRSA strains, with or without genes encoding Panton–Valentine leucocidin (PVL)—a toxin and key virulence factor of community-acquired strains—emerged in the 2000s [4]. Methicillin resistance is associated with the clinical outcomes of S. aureus infections [5], and the early treatment is important for a good prognosis of S. aureus bloodstream infections. The European Committee on Antimicrobial Susceptibility Testing recommends rapid antimicrobial susceptibility testing of S. aureus from positive blood culture bottles [6].
Methicillin resistance is conferred by mecA, which encodes penicillin-binding protein 2a. mecA is carried on the staphylococcal cassette chromosome mec (SCCmec), which is a mobile genetic element containing cassette chromosome recombinase genes (ccrA and ccrB) [7]. Although the transfer of SCCmec is a rare event in S. aureus, methicillin-susceptible S. aureus (MSSA) strains become MRSA after acquiring mecA embedded in SCCmec; all MRSA clones appear to harbor specific SCCmec types [8, 9].
SCCmec typing is based on the amplification of nine different genes (eight loci [A–H] of mec element sequences in addition to the mecA gene as an internal positive control), and subtypes are determined based on the presence or absence of various genes [10, 11]. The major SCCmec types in clinical isolates are types II, IV, and III. In contrast, MRSA SCCmec type V (SCCmec V), which has a novel ccrC gene [12], is infrequent among clinical isolates and has been isolated from humans and animals in Europe, Korea, Japan, Taiwan, and China. In Korea, SCCmec V strains have recently been isolated from patient serum samples and are considered an emerging pathogen [13]. Accordingly, microbiological laboratories in hospitals are monitoring the emergence of novel staphylococcal strains with antimicrobial resistance (AMR) in clinical settings.
Colonization or infection with livestock-associated MRSA has been reported in several domesticated livestock animals [14]. Carrier animals not only serve as reservoirs for opportunistic infections but can also transmit livestock-associated MRSA to other animal species or humans [15]. In Korea, livestock-associated SCCmec V strains have recently been isolated from pigs and their environments, posing serious public health concerns [16]. Based on the “One Health” concept, which is a comprehensive health control strategy for humans, animals, and the environment [17], circulating SCCmec V strains must be carefully monitored to maintain overall health.
Recently, we analyzed six SCCmec V isolates from a Korean University-affiliated hospital (Kangdong Sacred Heart Hospital, Hallym University College of Medicine) using whole-genome sequencing (WGS) to characterize the emerging SCCmec V strains. SCCmec can share genetic content via horizontal gene transfer (HGT) from MRSA or other methicillin-resistant staphylococci to MSSA or other methicillin-susceptible staphylococci among different hosts. SCCmec mobility relies on the function of ccr, encoding a protein that catalyzes host chromosome excision and insertion, which is a crucial step in HGT. However, few studies have evaluated the emergence or prevalence of SCCmec V in Korea.
We evaluated the possible phylogenetic emergence of SCCmec V via mecA HGT/direct transmission from overseas donor strains to Korean recipient strains through comparative genomic analysis of SCCmec V strains. To the best of our knowledge, this is the first study to estimate the emergence of SCCmec V in Korea and the donor–recipient relationship among overseas and Korean SCCmec V strains.
We screened the SCCmec types of MRSA strains deposited at Kangdong Sacred Heart Hospital (Seoul, Korea) from 2017 to 2020. We analyzed six SCCmec V strains isolated from patients with bacteremia or wound infections (Table 1). Ethical approval was not required as all data used in this study (host, strain, isolation date, isolation source, geographic location, and disease) related to SCCmec V strains were obtained from the publicly available National Center for Biotechnology Information (NCBI) database.
The presence of a gene encoding methicillin resistance was confirmed via PCR screening for mecA [10]. A single locus of repeat region X of the spa gene was sequenced and analyzed using Ridom StaphType v.3 (http://spaserver.ridom.de/) [18], and spa types were assigned using BioNumerics (v.7.5; Applied Math, Sint-Martens-Latem, Belgium). SCCmec was typed based on the mec and ccr gene complexes [19]. Genes encoding the PVL toxin were detected as described previously [4].
Six SCCmec V strains were inoculated onto 5% sheep blood agar plates and incubated at 35°C in the presence of 5% CO2 for 24 hrs. Single colonies were inoculated into broth cultures. Genomic DNA was extracted using the Blood and Cell Culture DNA Midi Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. For MiSeq sequencing (Illumina, San Diego, CA, USA), genomic libraries were prepared using the Nextera DNA Flex Library Prep Kit and sequenced on the Illumina MiSeq platform, as previously described [20]. Raw reads were processed using fastp v.0.20.0, with the default settings. For MinION sequencing (Oxford Nanopore Technologies, Oxford, UK), genomic libraries were constructed using a ligation sequencing kit (SQK-LSK109) according to the manufacturer’s instructions, and MinION flow cells (FlLO-MIN106D) were sequenced using MinION v.19.12.5.
For the MiSeq reads, adapter sequences were removed, and quality-filtering was performed using Trimmomatic v.0.39 [21]. For the MinION reads, base calling and demultiplexing were conducted using the Guppy GPU basecaller v.6.0.6, and sequencing artifacts were removed using Porechop v.0.2.4. Genome sequences were constructed from the MiSeq and MinION data using SPAdes v.3.14.0. Hybrid assembly was performed using Unicycler v.0.4.8, as previously described [20]. Genome assembly completeness was assessed using BUSCO v.5.3.2 with the lineage dataset bacillales_odb10. The complete genome sequences were annotated using the Prokaryotic Genome Annotation Pipeline v.4.3.
A genome map of the constructed MRSA genome was generated using CGView v.2.0.3. The CGView Comparison Tool (CCT) was used for visual comparison of the CGH comprising six WGS datasets, as described previously [22, 23]. All five other WGS datasets were aligned with the complete genome of strain HL24830 as a reference sequence. Clusters of orthologous gene (COG) functional categories were assigned based on the NCBI COG database (https://www.ncbi.nlm.nih.gov/research/cog) and are displayed using different colors.
Before conducting comparative genomic analysis, we retrieved all publicly available genome sequences of the genera Staphylococcus and Mammalicoccus assembled at the complete, chromosome, scaffold, and contig levels from the NCBI database. The sequences were reassigned for taxonomic classification based on the Genome Taxonomy Database (GTDB) using the GTDB-Tk toolkit v.2.0.0 before performing downstream analysis. SCCmec typing, multilocus sequence typing (MLST), and spa typing were performed using SCCmecFinder (https://bitbucket.org/genomicepidemiology/sccmecfinder.git), MLST (https://bitbucket.org/genomicepidemiology/mlst/src/master/), and spaTyper (https://bitbucket.org/genomicepidemiology/spatyper/src/main/), respectively. AMR and virulence-associated genes were identified via homology-based screening using AMRfinderPlus v.3.10.30 and the Virulence Factor Database (http://www.mgc.ac.cn/VFs/) [24], respectively. Genome sequences were annotated using Prokka v.1.14.6 to confirm whether the detected genes were functional. Using PorthoMCL, gene clusters were defined in the 513 MRSA genomes, and the clusters specific to each group were further analyzed.
Sixty-three complete circular genome sequences of SCCmec V MRSA with available information (host, strain, isolation date, isolation source, geographic location, and disease) were used to construct a phylogenetic tree. Using Snippy v.4.6.0, reference-based mapping was performed and single-nucleotide polymorphisms were identified. Recombination regions detected using Gubbins v.3.2.1 and the GTRGAMMA model were excluded. Based on recombination-free alignments, a maximum likelihood-based phylogenetic tree was constructed using RAxML v.8.2.11, with 1,000 bootstrap inferences. To estimate the emergence of the most common ancestors of the corresponding clades, Bayesian analysis was performed using BactDating v.1.1.
WGS datasets on 818 methicillin-resistant staphylococcal strains (including MRSA and other staphylococci) were retrieved from NCBI, and the strains were typed as SCCmec V using SCCmecFinder. Multiple sequence alignments of the mecA genes from these strains and the core sequences of the corresponding genomes were generated using Clustal Omega v.1.2.4. Based on the multiple alignments, a maximum likelihood-based phylogenetic tree was constructed using RAxML with 1,000 bootstrap inferences and the GTRGAMMA model. Macrococcus caseolyticus FDAARGOS_868 was used to root the tree, and the sequence was removed using Newick Utility v.1.6. We used Ranger-dtl 2.0 to estimate mecA HGT events among SCCmec V strains, using a support value cutoff of 0.9 to exclude ambiguous HGT events. The tree reconciliation method, which detects HGT events only when the overall genome-wide similarity of two strains is low compared to that of the target gene regions, thus excluding the direct transmission of MRSA strains with similar clonal backgrounds, was applied.
Table 2 presents the MinION and MiSeq sequencing metrics of the two assemblies from both WGS datasets. Table 3 summarizes the metrics of the six circularly assembled and annotated SCCmec V genomes. The complete genomes ranged from 2.809 to 2.926 Mbp, with a GC content of 32.5% in all datasets. Five genomes harbored one or two plasmids, and the six WGS datasets contained 2,626–2,790 protein-coding genes and 66–83 non-coding genes.
Fig. 1 shows the circular map of the six complete SCCmec V genomes (including the complete genome of HL24830 as a reference) generated using CGH. The CCT map consists of several circles showing the reference genome features and the results of Basic Local Alignment Search Tool comparisons between the reference and comparative sequences. The GC content and GC skew+/− were also evaluated. Several portions of the HL24830 genome sequence differed from the other five genomes, suggesting a unique trait of the former (Korean community-associated MRSA representative clone with ST72-t3092) compared with the other SCCmec V genomes.
Fig. 2 shows the dated phylogenetic tree of the six Korean MRSA SCCmec V strains and reported MRSA SCCmec V strains with complete genomes (N=57) obtained from NCBI. The dated tree showed five major clades. Strain NGA71 (isolated from human urine at the University of Benin Teaching Hospital, Nigeria) was an outlier in the tree. The German strains formed a major clade (clade 1), with a possible origin traced to the 1980s. Korean HL25870, HL25274, HL2018_N011-HL23187-HL20709, and HL24830 belonged to clades 1, 2, 3, and 4, respectively, suggesting diverse clades. A distinct clade comprised HL24830, which emerged in 2010 and was situated close to ER03364.3 (human outbreak strain isolated from blood at Mount Sinai Hospital, New York, NY, USA). Based on the dated findings, all six Korean SCCmec V strains may have emerged during 2000–2010.
Possible mecA HGT events in the SCCmec V recipient strains isolated in Korea are shown in Table 4. Six mecA HGT events with a support value of 1 were identified. We found an association between the Korean HL2018_N011 and HL20709 recipient strains and the P7 donor strain (a 2017 human outbreak strain from Hong Kong) and between the Korean HL25870 recipient strain and 11A731 donor strain (a 2011 Chinese food-origin strain). We also observed associations between the PCFA-221 recipient strain (a 2017 Korean pig isolate) and ISU 924 donor strain (a 2010 USA pig isolate) and between the BDH17 recipient strain (a 2017 Korean human isolate) and Sau55 donor strain (a 2006 human blood-origin strain from Taiwan).
MRSA strains with SCCmec V have been isolated from livestock and, less frequently, from humans. However, many people have companion animals in their homes, and medical hospitals and nursing homes have introduced animal-assisted therapy as a mental health service for patients and elderly residents [25]. Animals and humans are in constant close contact with the environment. Among bacterial pathogens with the potential to be transmitted between animals and humans, strains with AMR pose a serious threat to public health and must be closely monitored.
Staphylococcus pseudintermedius typically infects dogs. In 2008, mecA-positive S. pseudintermedius strains were recovered at two veterinary hospitals in Korea [26]. S. pseudintermedius with SCCmec V were the most prevalent isolates from veterinary staff (nose and hands), hospitalized animals (anus, skin, feet, ears, and wounds), and medical equipment (floors and phones). Pulsed-field gel electrophoresis analysis revealed genetic similarities among isolates from veterinary staff (hands), hospitalized animals (skin, feet, ears, and wounds), and medical equipment (floors) [26]. A recent study reported genetic relatedness in SCCmec between methicillin-resistant Staphylococcus isolates from companion dogs with pyoderma and their owners [27]. Among 31 dog–owner pairs, one pair with three isolates carrying SCCmec V, i.e., 18D20-1 (S. pseudintermedius, dog), 18D20-2 (S. schleiferi, dog), and 18H20-F2 (S. epidermidis, dog owner), was detected [27]. These observations suggest the clonal spread of SCCmec V strains between animals and humans.
Human invasive MRSA isolates harboring SCCmec V (N=3) were recovered at the Kangdong Sacred Heart Hospital between 2020 and 2021 [13]. A nosocomial outbreak of the community-associated MRSA P7 strain carrying SCCmec V−ST59 was reported in Hong Kong and originated from a clinical specimen from a neonatal intensive care unit [28]. We speculated that mecA was passed via HGT and/or direct transmission from the P7 donor strain (Hong Kong) to the HL2018_N011 and HL 20709 recipient strains (Korea) with ST59. The same ST59-t437 type was recovered in Hong Kong and Korea in 2018 [28], further supporting the notion of direct transmission occurring between these strains. However, HGT may have also played a role, as the computational procedure excluded the relatedness of similar clones, and we manually checked and denied the similarities of the complete genome sequences and P7 contigs. We observed mecA HGT from the 11A731 donor strain (China) to the HL25870 recipient strain (Korea) with ST1232. Chlebowicz, et al. [29] reported that SCCmec V from the MRSA strain UMCG-M4 was packaged into its bacteriophage capsids. Although transduction of complete SCCmec V was not observed, purified staphylococcal phage particles encapsulated large portions (including mecA) of SCCmec V. Therefore, it is necessary to evaluate the SCCmec V transduction/encapsulation capabilities of phages from the Korean recipient strains in the future.
The current study had two limitations. First, although mecA HGT events were estimated, possible HGT events associated with another SCCmec V component gene, ccrC, should be evaluated. Second, it remains unclear how SCCmec V of overseas donor strains from China, the USA, and Taiwan were transmitted to the Korean recipient strains. Possible routes include imported foods and immigrant animals or humans. Further studies monitoring the dynamics of SCCmec V clones in similar populations are required.
In conclusion, we estimated the possible clades and donor–recipient relationships among MRSA SCCmec V strains. We identified a unique Korean community-acquired MRSA SCCmec V strain harboring PVL-negative ST72-t3092, HL24830, and found that this strain differed from the other five Korean SCCmec V strains. The ST72 clone is prevalent among community-acquired MRSA isolates in Korea [30]. Continuous nationwide surveillance is required to monitor the emergence and spread of SCCmec V isolates in other Korean hospitals. The same clones with ST72-t3092-SCCmec V were recently isolated from a raw buffalo milk tank in Italy [30], suggesting that the risk of human MRSA colonization or infection may be associated with the handling of raw milk or consumption of contaminated dairy products.
SCCmec V of MRSA is an emerging clone in Korea and East Asian and European countries in which the V clone has been associated with human-to-animal or animal-to-human transmission. We expect that the investigation of this clone can enhance our understanding of the importance of “One Health.” The six Korean MRSA SCCmec V strains isolated from the blood or pus of patients may have emerged during 2000–2010. The clinical significance and implications of this clone among humans and pets/livestock/wild animals warrant further research to ensure that appropriate community health measures are taken. The silent AMR bacterial pandemic is an issue that requires prompt, optimized, and coordinated countermeasures.
Notes
AUTHOR CONTRIBUTIONS
Kim J-S and Takahashi T conceptualized the study. Kim H and Kim J-S were involved in the investigation. Kim H, Kim H-S, Kim HS, Song W, and Kim J-S were involved in the formal analysis. Kim H-S, Kim HS, Song W, and Kim J-S provided resources. Takahashi T and Kim J-S drafted the manuscript. Kim J-S, Kim H, and Takahashi T reviewed and edited the manuscript.
REFERENCES
1. Park SH. 1970; Antibiotic susceptibility of pathogenic microorganisms isolated in 1969. J Korean Med Assoc. 13:337–46.
2. Chong Y, Lee K. 2000; Present situation of antimicrobial resistance in Korea. J Infect Chemother. 6:189–95. DOI: 10.1007/s101560070001. PMID: 11810564.

3. Kim JS, Kim HS, Song W, Cho HC, Lee KM, Kim EC. 2007; Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolates with toxic shock syndrome toxin and staphylococcal enterotoxin C genes. Korean J Lab Med. 27:118–23. DOI: 10.3343/kjlm.2007.27.2.118. PMID: 18094562.
4. Lee SS, Kim YJ, Chung DR, Jung KS, Kim JS. 2010; Invasive infection caused by a community-associated methicillin-resistant Staphylococcus aureus strain not carrying Panton-Valentine leukocidin in South Korea. J Clin Microbiol. 48:311–3. DOI: 10.1128/JCM.00297-09. PMID: 19889903. PMCID: PMC2812302.

5. Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. 2003; Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 36:53–9. DOI: 10.1086/345476. PMID: 12491202.

6. Park JM, Kwon M, Hong KH, Lee H, Yong D. 2023; European Committee on Antimicrobial Susceptibility Testing-recommended rapid antimicrobial susceptibility testing of Escherichia coli, Klebsiella pneumoniae, and Staphylococcus aureus from positive blood culture bottles. Ann Lab Med. 43:443–50. DOI: 10.3343/alm.2023.43.5.443. PMID: 37080745. PMCID: PMC10151279.

7. Katayama Y, Ito T, Hiramatsu K. 2000; A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 44:1549–55. DOI: 10.1128/AAC.44.6.1549-1555.2000. PMID: 10817707. PMCID: PMC89911.

8. Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. 2002; The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A. 99:7687–92. DOI: 10.1073/pnas.122108599. PMID: 12032344. PMCID: PMC124322.
9. Oliveira DC, Tomasz A, de Lencastre H. 2002; Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect Dis. 2:180–9. DOI: 10.1016/S1473-3099(02)00227-X. PMID: 11944188.
10. Oliveira DC, de Lencastre H. 2002; Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 46:2155–61. DOI: 10.1128/AAC.46.7.2155-2161.2002. PMID: 12069968. PMCID: PMC127318.

11. Kim JS, Song W, Kim HS, Cho HC, Lee KM, Choi MS, et al. 2006; Association between the methicillin resistance of clinical isolates of Staphylococcus aureus, their staphylococcal cassette chromosome mec (SCCmec) subtype classification, and their toxin gene profiles. Diagn Microbiol Infect Dis. 56:289–95. DOI: 10.1016/j.diagmicrobio.2006.05.003. PMID: 16854552.
12. Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. 2004; Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother. 48:2637–51. DOI: 10.1128/AAC.48.7.2637-2651.2004. PMID: 15215121. PMCID: PMC434217.

13. Lade H, Chung SH, Lee Y, Joo HS, Kim JS. 2022; Genotypes of Staphylococcus aureus clinical isolates are associated with phenol-soluble modulin (PSM) production. Toxins (Basel). 14:556. DOI: 10.3390/toxins14080556. PMID: 36006218. PMCID: PMC9412541.

14. Aires-de-Sousa M. 2017; Methicillin-resistant Staphylococcus aureus among animals: current overview. Clin Microbiol Infect. 23:373–80. DOI: 10.1016/j.cmi.2016.11.002. PMID: 27851997.
15. Wang Y, Zhang P, Wu J, Chen S, Jin Y, Long J, et al. Transmission of livestock-associated methicillin-resistant Staphylococcus aureus between animals, environment, and humans in the farm. Environ Sci Pollut Res Int. doi: 10.1007/s11356-023-28532-7. Epub ahead of print. DOI: 10.1007/s11356-023-28532-7. PMID: 37418185.
16. Back SH, Eom HS, Lee HH, Lee GY, Park KT, Yang SJ. 2020; Livestock-associated methicillin-resistant Staphylococcus aureus in Korea: antimicrobial resistance and molecular characteristics of LA-MRSA strains isolated from pigs, pig farmers, and farm environment. J Vet Sci. 21:e2. DOI: 10.4142/jvs.2020.21.e2. PMID: 31940681. PMCID: PMC7000904.

17. Centers for Disease Control and Prevention. One Health. https://www.cdc.gov/onehealth/index.html. Updated on July 2023.
18. Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, et al. 2003; Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 41:5442–8. DOI: 10.1128/JCM.41.12.5442-5448.2003. PMID: 14662923. PMCID: PMC309029.

19. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, et al. 2007; Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 51:264–74. DOI: 10.1128/AAC.00165-06. PMID: 17043114. PMCID: PMC1797693.

20. Kim JS, Sakaguchi S, Fukushima Y, Yoshida H, Nakano T, Takahashi T. 2020; Complete genome sequences of four Streptococcus canis strains isolated from dogs in South Korea. Microbiol Resour Announc. 9:e00818–20. DOI: 10.1128/MRA.00818-20. PMID: 32817161. PMCID: PMC7427199.
21. Bolger AM, Lohse M, Usadel B. 2014; Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–20. DOI: 10.1093/bioinformatics/btu170. PMID: 24695404. PMCID: PMC4103590.

22. Grant JR, Arantes AS, Stothard P. 2012; Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genomics. 13:202. DOI: 10.1186/1471-2164-13-202. PMID: 22621371. PMCID: PMC3469350.

23. Kim JM, Fukushima Y, Yoshida H, Kim JS, Takahashi T. 2022; Comparative genomic features of Streptococcus canis based on pan-genome orthologous group analysis according to sequence type. Jpn J Infect Dis. 75:269–76. DOI: 10.7883/yoken.JJID.2021.533. PMID: 34588372.

24. Yoshida H, Kim JM, Maeda T, Goto M, Tsuyuki Y, Shibata S, et al. 2023; Virulence-associated genome sequences of Pasteurella canis and unique toxin gene prevalence of P. canis and Pasteurella multocida isolated from humans and companion animals. Ann Lab Med. 43:263–72. DOI: 10.3343/alm.2023.43.3.263. PMID: 36544338. PMCID: PMC9791007.

25. Thodberg K, Videbech PB, Hansen TGB, Pedersen AB, Christensen JW. 2021; Dog visits in nursing homes - increase complexity or keep it simple? A randomised controlled study. PLoS One. 16:e0251571. DOI: 10.1371/journal.pone.0251571. PMID: 34038451. PMCID: PMC8153477.

26. Moon BY, Youn JH, Shin S, Hwang SY, Park YH. 2012; Genetic and phenotypic characterization of methicillin-resistant staphylococci isolated from veterinary hospitals in South Korea. J Vet Diagn Invest. 24:489–98. DOI: 10.1177/1040638712440985. PMID: 22529115.

27. Kang JH, Hwang CY. 2021; One health approach to genetic relatedness in SCCmec between methicillin-resistant Staphylococcus isolates from companion dogs with pyoderma and their owners. Vet Microbiol. 253:108957. DOI: 10.1016/j.vetmic.2020.108957. PMID: 33385887.
28. Cheng VCC, Wong SC, Cao H, Chen JHK, So SYC, Wong SCY, et al. 2019; Whole-genome sequencing data-based modeling for the investigation of an outbreak of community-associated methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit in Hong Kong. Eur J Clin Microbiol Infect Dis. 38:563–73. DOI: 10.1007/s10096-018-03458-y. PMID: 30680562.

29. Chlebowicz MA, Mašlaňová I, Kuntová L, Grundmann H, Pantůček R, Doškař J, et al. 2014; The staphylococcal cassette chromosome mec type V from Staphylococcus aureus ST398 is packaged into bacteriophage capsids. Int J Med Microbiol. 304:764–74. DOI: 10.1016/j.ijmm.2014.05.010. PMID: 24951306.
30. Giovanni N, Elisa S, Marta C, Rosa F, Loredana C, Alessandra B, et al. 2020; Occurrence and characteristics of methicillin-resistant Staphylococcus aureus (MRSA) in buffalo bulk tank milk and the farm workers in Italy. Food Microbiol. 91:103509. DOI: 10.1016/j.fm.2020.103509. PMID: 32539967.
Fig. 1
Circular map of the six Korean complete MRSA SCCmec V strain genomes (including the HL24830 reference strain). A genome map of the constructed MRSA genome was generated using CGView v.2.0.3. The CGView Comparison Tool was used to construct the circular plot of Staphylococcus aureus DNA sequences. COG features for protein-coding genes and locations of protein-coding, transfer RNA, and ribosomal RNA genes are presented on the four outermost concentric rings. Similar regions of MRSA SCCmec V strains are presented on the next six concentric rings. Based on the percent identity determined using the Basic Local Alignment Search Tool between the similar regions, the inside rings of the MRSA genomes appear black (100% identical) to blue (≥88% identical). The black peaks depict GC content, and the innermost green and purple peaks describe positive and negative GC skews, respectively.
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; COG, clusters of orthologous gene; SCCmec V, staphylococcal cassette chromosome mec type V.
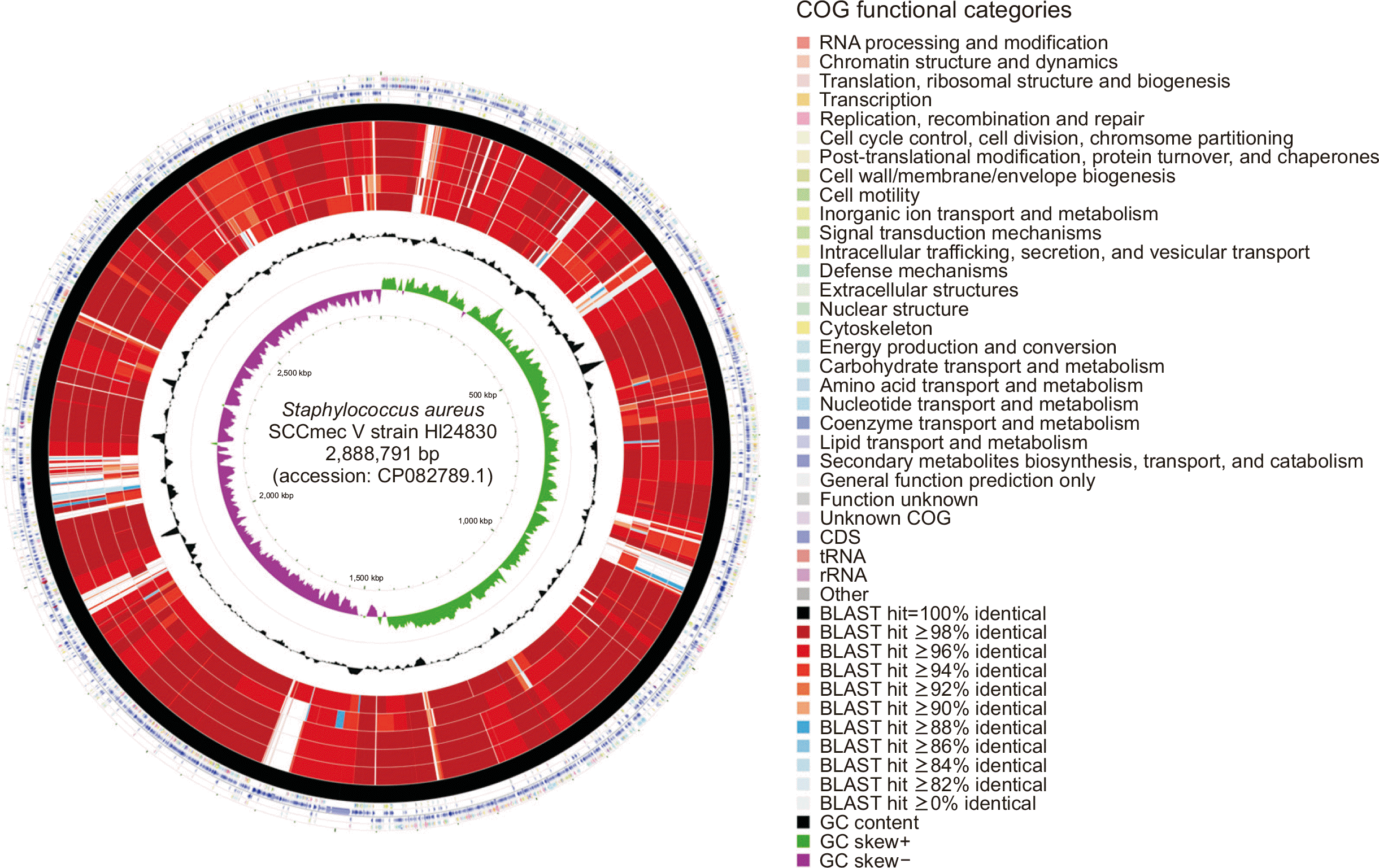
Fig. 2
Dated phylogenetic tree, including the six Korean MRSA SCCmec V strains and other MRSA SCCmec V strains with complete genomes (N= 57) available from the National Center for Biotechnology Information database. Strain HL24830 appears around 2010 and is situated near strain ER03364.3 from the USA. The six Korean SCCmec V stains may have emerged during 2000–2010.
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; SCCmec V, staphylococcal cassette chromosome mec type V.
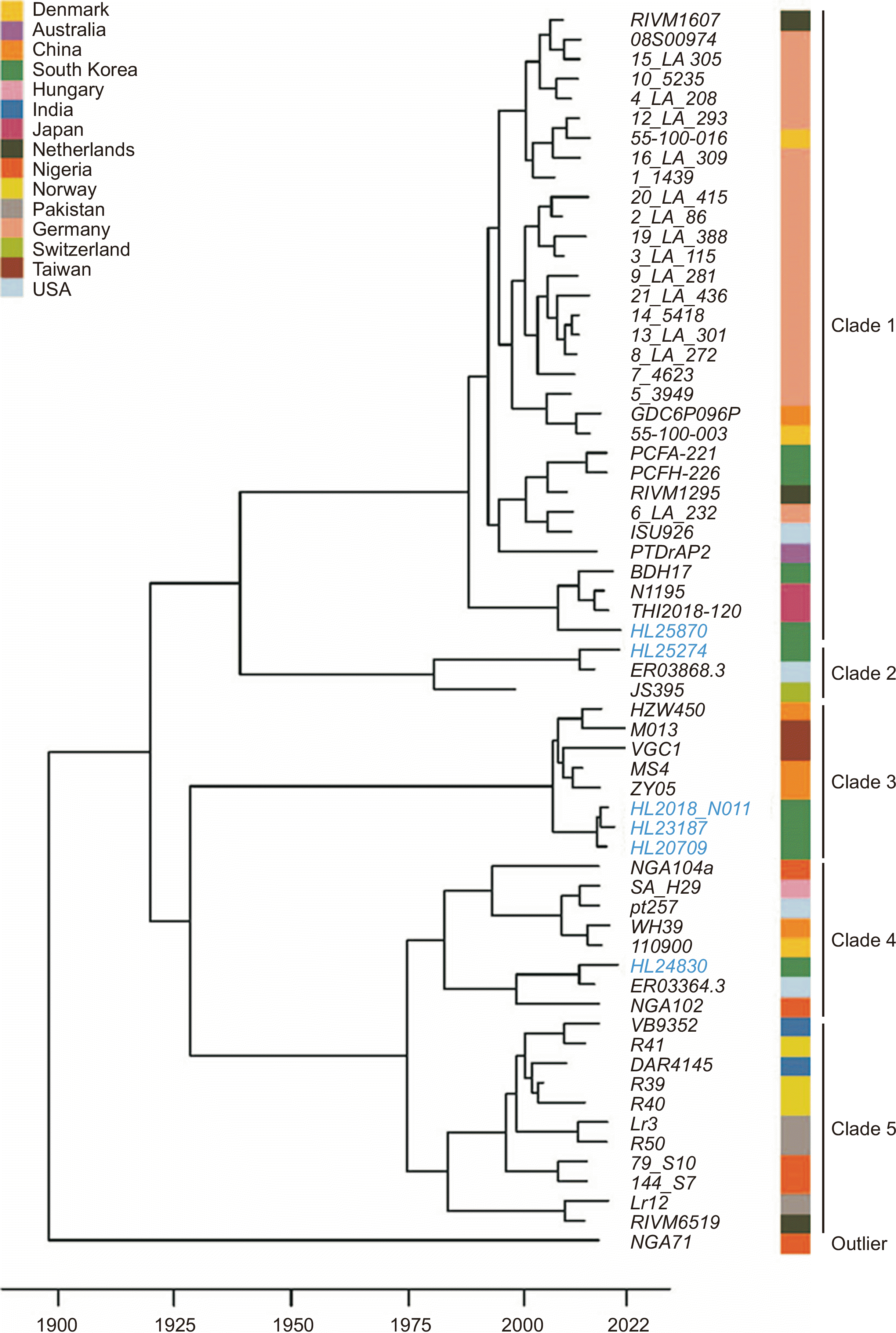
Table 1
Strain information
Table 2
Sequencing metrics for the two assemblies from two whole-genome sequencing datasets
| Strain | MinION sequencing* | MiSeq sequencing† | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Read length N50 (bp) | Mean read quality | N of reads | Total bases (bp) | Coverage (X) | N of reads | Total bases (bp) | Coverage (X) | ||||
| HL20709 | 13,266 | 11.3 | 112,634 | 1,220,990,317 | 426.9 | 1,947,068 | 288,474,163 | 100.9 | |||
| HL23187 | 16,867 | 11.2 | 42,440 | 557,671,576 | 196.4 | 1,837,608 | 272,537,071 | 96.0 | |||
| HL25274 | 3,247 | 12.6 | 329,151 | 762,618,177 | 271.4 | 3,027,274 | 445,604,778 | 158.6 | |||
| HL24830 | 7,148 | 8.7 | 132,592 | 528,929,395 | 181.5 | 1,881,702 | 278,662,641 | 95.6 | |||
| HL25870 | 7,928 | 8.7 | 335,981 | 1,402,136,279 | 479.1 | 1,720,578 | 255,003,766 | 87.1 | |||
| HL2018_N011 | 8,616 | 8.7 | 81,207 | 325,914,573 | 116.0 | 1,613,702 | 238,922,352 | 85.0 | |||
Table 3
Summarized metrics for the six circularly assembled and annotated genomes
| Metric | HL20709 | HL23187 | HL25274 | HL24830 | HL25870 | HL2018_N011 |
|---|---|---|---|---|---|---|
| Completeness, %* | 450 (100%) | 450 (100%) | 450 (100%) | 450 (100%) | 450 (100%) | 450 (100%) |
| N of chromosomes | 1 | 1 | 1 | 1 | 1 | 1 |
| N of plasmids | 0 | 1 | 2 | 1 | 1 | 1 |
| Total length, bp | 2,860,303 | 2,860,005 | 2,913,602 | 2,926,847 | 2,809,575 | 2,881,027 |
| GC, % | 32.5 | 32.5 | 32.5 | 32.5 | 32.5 | 32.5 |
| N of protein-coding genes | 2,706 | 2,695 | 2,714 | 2,790 | 2,626 | 2,705 |
| N of non-coding genes | 66 | 82 | 83 | 82 | 82 | 81 |
Table 4
Computational analysis of possible mecA HGT in staphylococcal SCCmec type V strains isolated in Korea
| Donor strain | Host | Collection year | Oversea country | Recipient strain | Host (specimen) | Collection year | Location | Support value |
|---|---|---|---|---|---|---|---|---|
| S. aureus ISU 924 | Pig | 2010 | The USA | S. aureus PCFA-221 | Pig (nasal cavity) | 2017 | Chungcheongnam-do | 1 |
| S. aureus Sau55, 5_3949, VET0107R, Sau88, RR17, W, VET0421R, CFSA16SA028, CFSA15SA094, 08-01728, VET0241R, and VET0176R | Human | 2006 | Taiwan, Germany, and the Netherlands | S. aureus BDH17 | Human | 2019 | Unknown location | 1 |
| S. aureus P7* | Human (pus) | 2017 | Hong Kong | S. aureus HL2018_N011 | Human (pus) | 2018 | Seoul | 1 |
| S. aureus P7* | Human (pus) | 2017 | Hong Kong | S. aureus HL20709 | Human (blood) | 2017 | Seoul | 1 |
| S. aureus VET0077R | Human | 2008 | Unknown | S. schleiferi OT1-1 | Dog (ear) | 2017 | Seongnam-si | 1 |
| S. aureus 11A731 | Food | 2011 | China | S. aureus HL25870 | Human (blood) | 2020 | Seoul | 1 |
| S. epidermidis Se_BPH0736 and HD43-1 | Human (nasal cavity and prosthetic joint) | 2011 and 2018 | Australia and Germany | S. epidermidis CDC121 | Human (skin) | 2017 | Seoul | 0.92 |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download