1. Weiner-Lastinger LM, Abner S, Edwards JR, Kallen AJ, Karlsson M, Magill SS, et al. Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: summary of data reported to the National Healthcare Safety Network, 2015-2017. Infect Control Hosp Epidemiol. 2020; 41:1–18. DOI:
10.1017/ice.2019.296. PMID:
31767041. PMCID:
PMC8276252.

2. Lucena A, Dalla Costa LM, Nogueira KS, Matos AP, Gales AC, Paganini MC, et al. 2014; Nosocomial infections with metallo-beta-lactamase-producing
Pseudomonas aeruginosa: molecular epidemiology, risk factors, clinical features and outcomes. J Hosp Infect. 87:234–40. DOI:
10.1016/j.jhin.2014.05.007. PMID:
25027563.
3. Diene SM, Rolain JM. 2014; Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 20:831–8. DOI:
10.1111/1469-0691.12655. PMID:
24766097.

5. Almarzoky Abuhussain SS, Sutherland CA, Nicolau DP. 2019; In vitro potency of antipseudomonal beta-lactams against blood and respiratory isolates of
P. aeruginosa collected from US hospitals. J Thorac Dis. 11:1896–902. DOI:
10.21037/jtd.2019.05.13. PMID:
31285882. PMCID:
PMC6588753.
6. Woodworth KR, Walters MS, Weiner LM, Edwards J, Brown AC, Huang JY, et al. Vital signs: containment of novel multidrug-resistant organisms and resistance mechanisms - United States, 2006-2017. MMWR Morb Mortal Wkly Rep. 2018; 67:396–401. DOI:
10.15585/mmwr.mm6713e1. PMID:
29621209. PMCID:
PMC5889247.

7. Yong D, Shin HB, Kim YK, Cho J, Lee WG, Ha GY, et al. 2014; Increase in the prevalence of carbapenem-resistant
Acinetobacter isolates and ampicillin-resistant non-typhoidal
Salmonella species in Korea: a KONSAR study conducted in 2011. Infect Chemother. 46:84–93. DOI:
10.3947/ic.2014.46.2.84. PMID:
25024870. PMCID:
PMC4091365.

8. Bae MH, Kim MS, Kim TS, Kim S, Yong D, Ha GY, et al. 2023; Changing epidemiology of pathogenic bacteria over the past 20 years in Korea. J Korean Med Sci. 38:e73. DOI:
10.3346/jkms.2023.38.e73. PMID:
36918027. PMCID:
PMC10010907.

9. Wang MG, Liu ZY, Liao XP, Sun RY, Li RB, Liu Y, et al. 2021; Retrospective data insight into the global distribution of carbapenemase-producing
Pseudomonas aeruginosa. Antibiotics (Basel). 10:548. DOI:
10.3390/antibiotics10050548. PMID:
34065054. PMCID:
PMC8151531.

10. Livermore DM. 2002; Multiple mechanisms of antimicrobial resistance in
Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis. 34:634–40. DOI:
10.1086/338782. PMID:
11823954.
12. Grupper M, Sutherland C, Nicolau DP. 2017; Multicenter evaluation of ceftazidime-avibactam and ceftolozane-tazobactam inhibitory activity against meropenem-nonsusceptible
Pseudomonas aeruginosa from blood, respiratory tract, and wounds. Antimicrob Agents Chemother. 61:e00875–17. DOI:
10.1128/AAC.00875-17. PMID:
28739780. PMCID:
PMC5610483.
13. Zavascki AP, Barth AL, Gonçalves AL, Moro AL, Fernandes JF, Martins AF, et al. 2006; The influence of metallo-beta-lactamase production on mortality in nosocomial
Pseudomonas aeruginosa infections. J Antimicrob Chemother. 58:387–92. DOI:
10.1093/jac/dkl239. PMID:
16751638.
14. Corvec S, Poirel L, Espaze E, Giraudeau C, Drugeon H, Nordmann P. 2008; Long-term evolution of a nosocomial outbreak of
Pseudomonas aeruginosa producing VIM-2 metallo-enzyme. J Hosp Infect. 68:73–82. DOI:
10.1016/j.jhin.2007.10.016. PMID:
18079018.

15. Kouda S, Ohara M, Onodera M, Fujiue Y, Sasaki M, Kohara T, et al. 2009; Increased prevalence and clonal dissemination of multidrug-resistant
Pseudomonas aeruginosa with the
blaIMP-1 gene cassette in Hiroshima. J Antimicrob Chemother. 64:46–51. DOI:
10.1093/jac/dkp142. PMID:
19398456.

16. CLSI. 2021. Performance standards for antimicrobial susceptibility testing. 31st informational supplement. CLSI M100-S31. Clinical and Laboratory Standards Institute;Wayne, PA:
17. Jeong S, Lee N, Park MJ, Jeon K, Kim HS, Kim HS, et al. 2022; Genotypic distribution and antimicrobial susceptibilities of carbapenemase-producing
Enterobacteriaceae isolated from rectal and clinical samples in Korean university hospitals between 2016 and 2019. Ann Lab Med. 42:36–46. DOI:
10.3343/alm.2022.42.1.36. PMID:
34374347. PMCID:
PMC8368229.

18. Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. 1988; CDC definitions for nosocomial infections, 1988. Am J Infect Control. 16:128–40. DOI:
10.1016/0196-6553(88)90053-3. PMID:
2841893.

19. Bongiovanni M, Barilaro G, Zanini U, Giuliani G. 2022; Impact of the COVID-19 pandemic on multidrug-resistant hospital-acquired bacterial infections. J Hosp Infect. 123:191–2. DOI:
10.1016/j.jhin.2022.02.015. PMID:
35245646. PMCID:
PMC8885440.

20. Jeon K, Jeong S, Lee N, Park MJ, Song W, Kim HS, et al. 2022; Impact of COVID-19 on antimicrobial consumption and spread of multidrug-resistance in bacterial infections. Antibiotics (Basel). 11:535. DOI:
10.3390/antibiotics11040535. PMID:
35453286. PMCID:
PMC9025690.

23. Perez LRR, Carniel E, Narvaez GA. 2022; Emergence of NDM-producing
Pseudomonas aeruginosa among hospitalized patients and impact on antimicrobial therapy during the coronavirus disease 2019 (COVID-19) pandemic. Infect Control Hosp Epidemiol. 43:1279–80. DOI:
10.1017/ice.2021.253. PMID:
34184628.
24. Lee K, Lim JB, Yum JH, Yong D, Chong Y, Kim JM, et al. 2002; bla(VIM-2) cassette-containing novel integrons in metallo-beta-lactamase-producing
Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob Agents Chemother. 46:1053–8. DOI:
10.1128/AAC.46.4.1053-1058.2002. PMID:
11897589. PMCID:
PMC127086.

25. Seok Y, Bae IK, Jeong SH, Kim SH, Lee H, Lee K. 2011; Dissemination of IMP-6 metallo-beta-lactamase-producing
Pseudomonas aeruginosa sequence type 235 in Korea. J Antimicrob Chemother. 66:2791–6. DOI:
10.1093/jac/dkr381. PMID:
21933788.
26. Park Y, Koo SH. 2022; Epidemiology, molecular characteristics, and virulence factors of carbapenem-resistant
Pseudomonas aeruginosa isolated from patients with urinary tract infections. Infect Drug Resist. 15:141–51. DOI:
10.2147/IDR.S346313. PMID:
35058697. PMCID:
PMC8765443.
27. Hong JS, Song W, Park MJ, Jeong S, Lee N, Jeong SH. 2021; Molecular characterization of the first emerged NDM-1-producing
Pseudomonas aeruginosa isolates in South Korea. Microb Drug Resist. 27:1063–70. DOI:
10.1089/mdr.2020.0374. PMID:
33332204.
28. Khan A, Shropshire WC, Hanson B, Dinh AQ, Wanger A, Ostrosky-Zeichner L, et al. 2020; Simultaneous infection with
Enterobacteriaceae and
Pseudomonas aeruginosa harboring multiple carbapenemases in a returning traveler colonized with Candida auris. Antimicrob Agents Chemother. 64:e01466–19. DOI:
10.1128/AAC.01466-19. PMID:
31658962. PMCID:
PMC6985746.
29. Perez F, El Chakhtoura NG, Papp-Wallace KM, Wilson BM, Bonomo RA. 2016; Treatment options for infections caused by carbapenem-resistant Enterobacteriaceae: can we apply "precision medicine" to antimicrobial chemotherapy? Expert Opin Pharmacother. 17:761–81. DOI:
10.1517/14656566.2016.1145658. PMID:
26799840. PMCID:
PMC4970584.
30. Marshall S, Hujer AM, Rojas LJ, Papp-Wallace KM, Humphries RM, Spellberg B, et al. 2017; Can ceftazidime-avibactam and aztreonam overcome β-lactam resistance conferred by metallo-β-lactamases in
Enterobacteriaceae? Antimicrob Agents Chemother. 61:e02243–16. DOI:
10.1128/AAC.02243-16. PMID:
28167541. PMCID:
PMC5365724.
32. Esposito F, Cardoso B, Fontana H, Fuga B, Cardenas-Arias A, Moura Q, et al. 2021; Genomic analysis of carbapenem-resistant
Pseudomonas aeruginosa isolated from urban rivers confirms spread of clone sequence Type 277 carrying broad resistome and virulome beyond the hospital. Front Microbiol. 12:701921. DOI:
10.3389/fmicb.2021.701921. PMID:
34539602. PMCID:
PMC8446631.
33. Mauri C, Maraolo AE, Di Bella S, Luzzaro F, Principe L. 2021; The revival of aztreonam in combination with avibactam against metallo-β-lactamase-producing gram-negatives: a systematic review of in vitro studies and clinical cases. Antibiotics (Basel). 10:1012. DOI:
10.3390/antibiotics10081012. PMID:
34439062. PMCID:
PMC8388901.

34. Barron MA, Richardson K, Jeffres M, McCollister B. 2016; Risk factors and influence of carbapenem exposure on the development of carbapenem resistant
Pseudomonas aeruginosa bloodstream infections and infections at sterile sites. Springerplus. 5:755. DOI:
10.1186/s40064-016-2438-4. PMID:
27386239. PMCID:
PMC4912523.
35. Tsao LH, Hsin CY, Liu HY, Chuang HC, Chen LY, Lee YJ. 2018; Risk factors for healthcare-associated infection caused by carbapenem-resistant
Pseudomonas aeruginosa. J Microbiol Immunol Infect. 51:359–66. DOI:
10.1016/j.jmii.2017.08.015. PMID:
28988663.
37. Hirakata Y, Yamaguchi T, Nakano M, Izumikawa K, Mine M, Aoki S, et al. 2003; Clinical and bacteriological characteristics of IMP-type metallo-beta-lactamase-producing
Pseudomonas aeruginosa. Clin Infect Dis. 37:26–32. DOI:
10.1086/375594. PMID:
12830405.

38. Shi Q, Huang C, Xiao T, Wu Z, Xiao Y. 2019; A retrospective analysis of
Pseudomonas aeruginosa bloodstream infections: prevalence, risk factors, and outcome in carbapenem-susceptible and -non-susceptible infections. Antimicrob Resist Infect Control. 8:68. DOI:
10.1186/s13756-019-0520-8. PMID:
31057792. PMCID:
PMC6485151.
39. Zhang Y, Li Y, Zeng J, Chang Y, Han S, Zhao J, et al. 2020; Risk factors for mortality of inpatients with
Pseudomonas aeruginosa bacteremia in China: impact of resistance profile in the mortality. Infect Drug Resist. 13:4115–23. DOI:
10.2147/IDR.S268744. PMID:
33209041. PMCID:
PMC7669529.
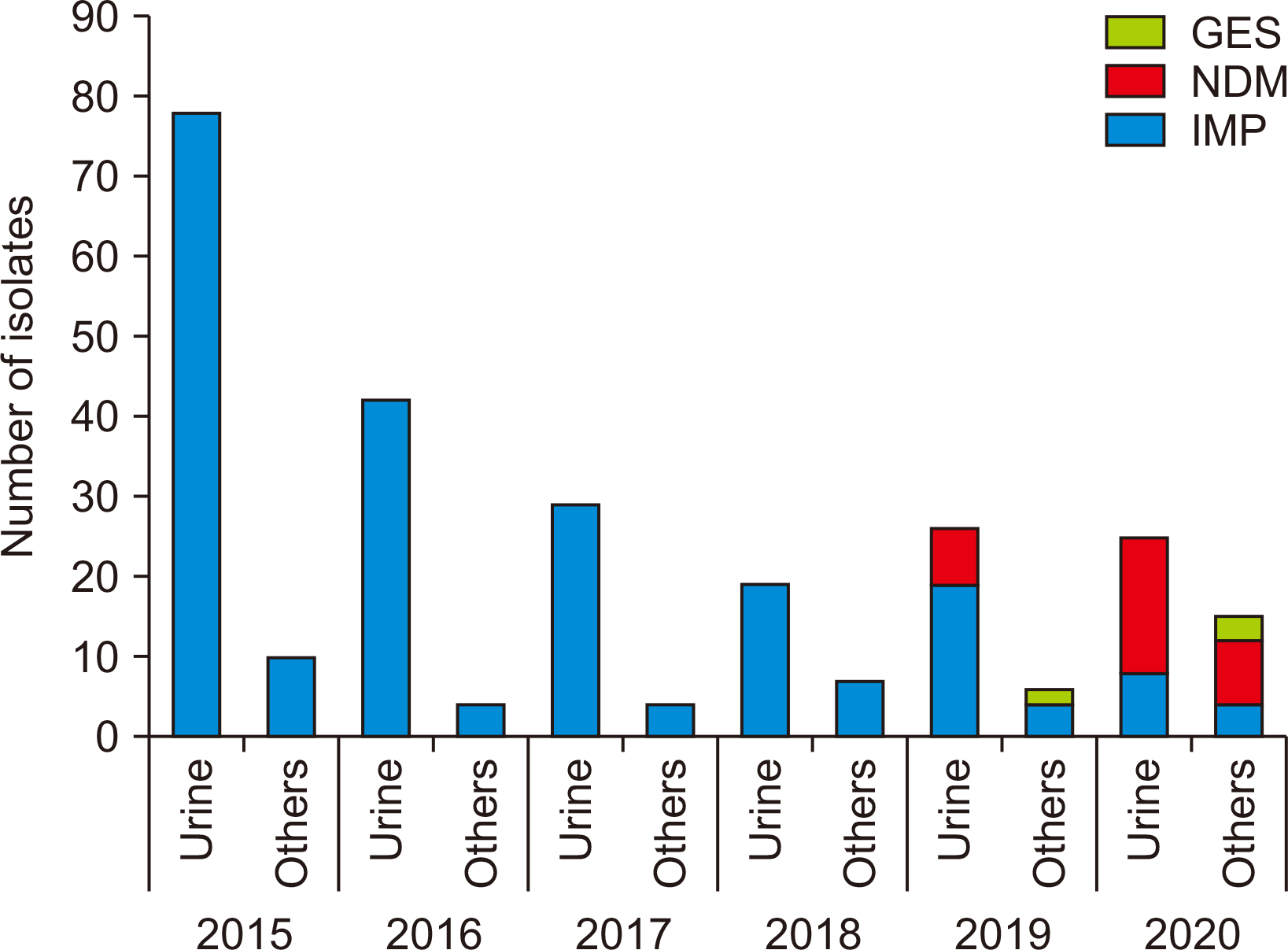
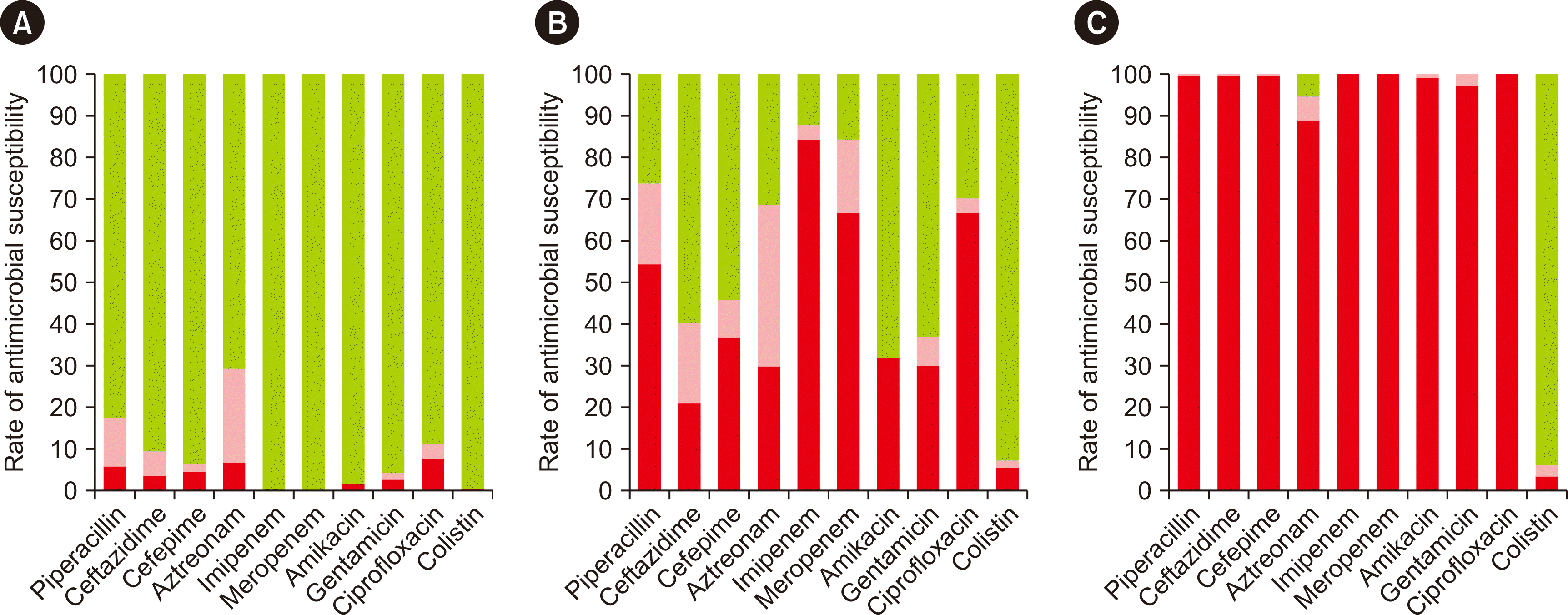
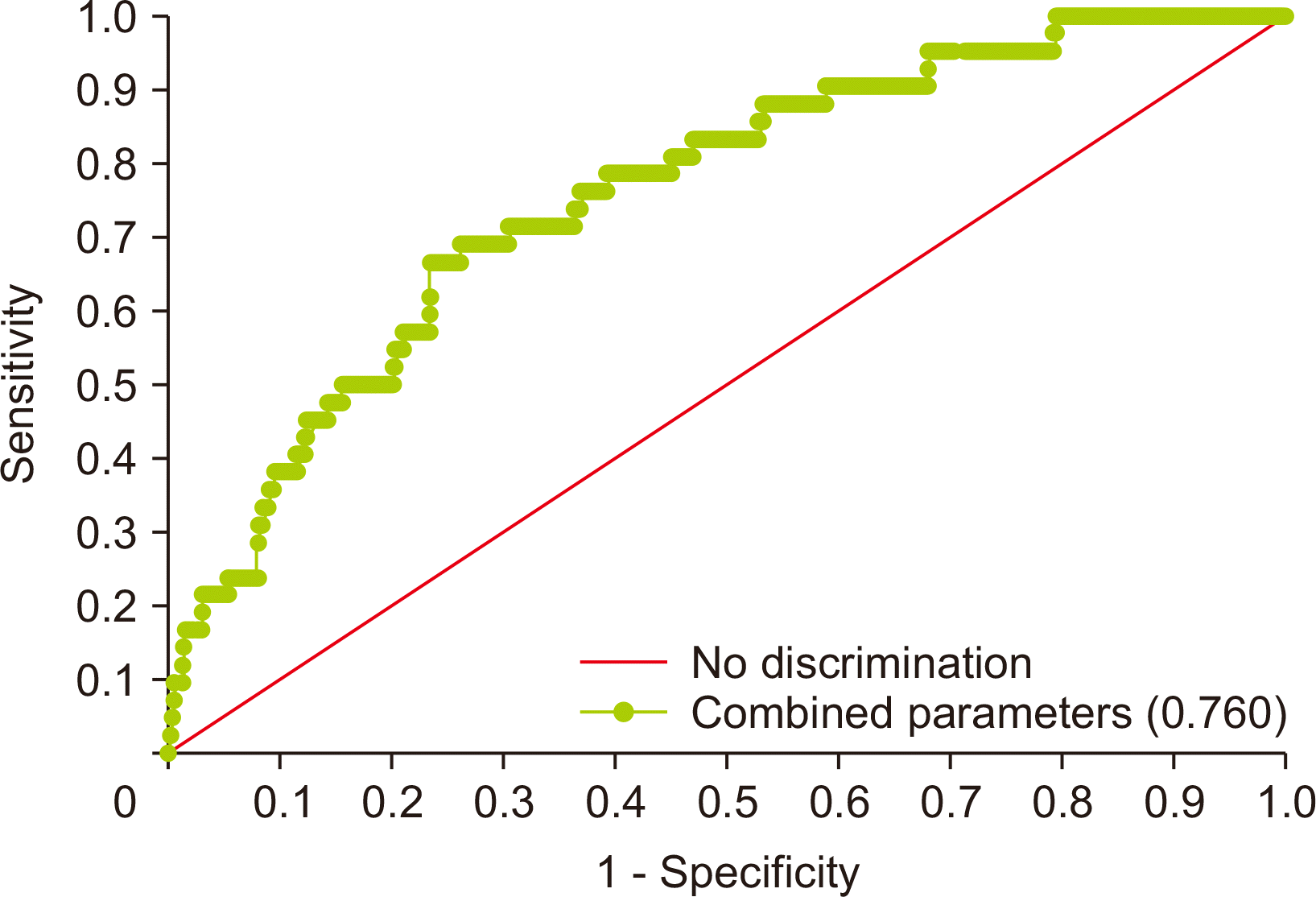




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download