1. Yoon HK, Yang HL, Jung CW, Lee HC. Artificial intelligence in perioperative medicine: a narrative review. Korean J Anesthesiol. 2022; 75:202–15.

2. Sessler DI. Big Data–and its contributions to perioperative medicine. Anaesthesia. 2014; 69:100–5.

4. Sjoding MW, Dickson RP, Iwashyna TJ, Gay SE, Valley TS. Racial bias in pulse oximetry measurement. N Engl J Med. 2020; 383:2477–8.

5. Komorowski M, Celi LA, Badawi O, Gordon AC, Faisal AA. The Artificial Intelligence Clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med. 2018; 24:1716–20.

6. Jiang LY, Liu XC, Nejatian NP, Nasir-Moin M, Wang D, Abidin A, et al. Health system-scale language models are all-purpose prediction engines. Nature;2023. doi: 10.1038/s41586-023-06160-y. [Epub ahead of print].
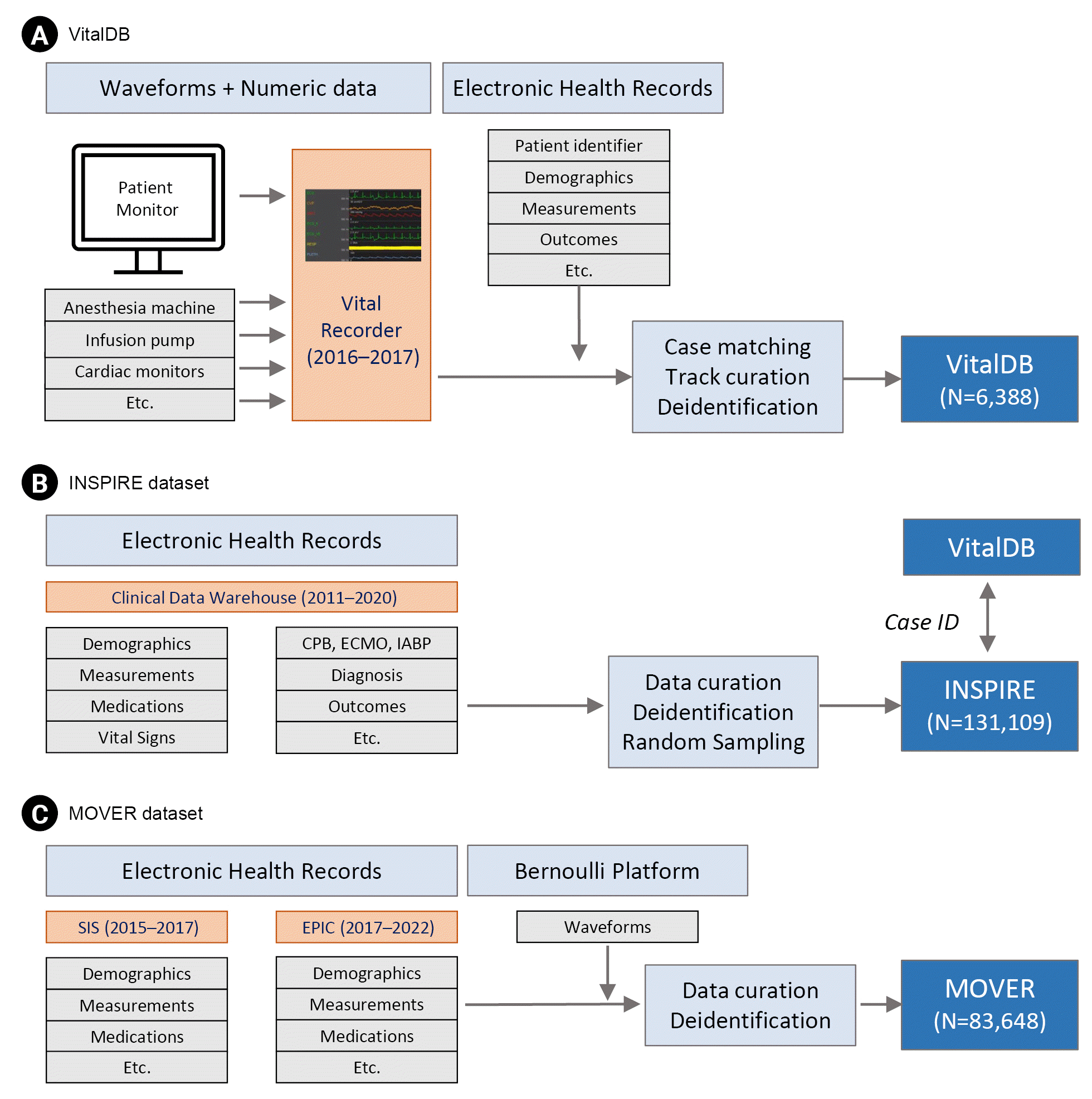
7. Lee HC, Park Y, Yoon SB, Yang SM, Park D, Jung CW. VitalDB, a high-fidelity multi-parameter vital signs database in surgical patients. Sci Data. 2022; 9:279.

8. Lee HC, Jung CW. Vital Recorder-a free research tool for automatic recording of high-resolution time-synchronized physiological data from multiple anaesthesia devices. Sci Rep. 2018; 8:1527.
9. Moody GB, Mark RG, Goldberger AL. PhysioNet: a research resource for studies of complex physiologic and biomedical signals. Comput Cardiol. 2000; 27:179–82.

10. Samad M, Rinehart J, Angel M, Kanomata Y, Baldi P, Cannesson M. MOVER: Medical Informatics Operating Room Vitals and Events Repository. medRxiv. 2023; 2023.03.03.23286777.

11. Sauer CM, Dam TA, Celi LA, Faltys M, De La Hoz MAA, Adhikari L, et al. Systematic review and comparison of publicly available ICU aata sets-a decision guide for clinicians and data scientists. Crit Care Med. 2022; 50:e581–8.
12. Moody GB, Mark RG. The impact of the MIT-BIH arrhythmia database. IEEE Eng Med Biol Mag. 2001; 20:45–50.

13. Johnson AEW, Pollard TJ, Shen L, Lehman LWH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016; 3:160035.

14. Johnson AEW, Bulgarelli L, Shen L, Gayles A, Shammout A, Horng S, et al. MIMIC-IV, a freely accessible electronic health record dataset. Sci Data. 2023; 10:1.

15. Johnson AEW, Pollard TJ, Berkowitz SJ, Greenbaum NR, Lungren MP, et al. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci Data. 2019; 6:317.

16. Pollard TJ, Johnson AEW, Raffa JD, Celi LA, Mark RG, Badawi O. The eICU Collaborative Research Database, a freely available multicenter database for critical care research. Sci Data. 2018; 5:180178.

17. Thoral PJ, Peppink JM, Driessen RH, Sijbrands EJG, Kompanje EJO, Kaplan L, et al. Sharing ICU Patient Data Responsibly Under the Society of Critical Care Medicine/European Society of Intensive Care Medicine Joint Data Science Collaboration: The Amsterdam University Medical Centers Database (AmsterdamUMCdb) Example. Crit Care Med. 2021; 49:e563–77.

18. Hyland SL, Faltys M, Hüser M, Lyu X, Gumbsch T, Esteban C, et al. Early prediction of circulatory failure in the intensive care unit using machine learning. Nat Med. 2020; 26:364–73.

19. Stupple A, Singerman D, Celi LA. The reproducibility crisis in the age of digital medicine. NPJ Digit Med. 2019; 2:2.





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download