This article has been
cited by other articles in ScienceCentral.
Abstract
Glutathione (GSH) is a chief cellular antioxidant, affecting stem cell functions. The cellular GSH level is dynamically altered by the redox buffering system and transcription factors, including NRF2. Additionally, GSH is differentially regulated in each organelle. We previously reported a protocol for monitoring the real-time GSH levels in live stem cells using the reversible GSH sensor FreSHtracer. However, GSH-based stem cell analysis needs be comprehensive and organelle-specific. Hence, in this study, we demonstrate a detailed protocol to measure the GSH regeneration capacity (GRC) in living stem cells by measuring the intensities of the FreSHtracer and the mitochondrial GSH sensor MitoFreSHtracer using a high-content screening confocal microscope. This protocol typically analyses the GRC in approximately 4 h following the seeding of the cells onto plates. This protocol is simple and quantitative. With some minor modifications, it can be employed flexibly to measure the GRC for the whole-cell area or just the mitochondria in all adherent mammalian stem cells.
Keywords: Glutathione, Stem cells, FreSHtracer, Glutathione regeneration capacity
Introduction
Glutathione (GSH) is a major thiol-based cellular antioxidant that indicates cellular redox equilibrium (
1,
2). Recently, we reported that GSH levels determine stem cell functions and therapeutic efficacy in asthma and graft-versus-host diseases (
3-
5). Thus, monitoring GSH levels is crucial for the development of therapeutic stem cells. However, cellular GSH levels change dynamically based on culture conditions such as confluence and passage number (
3). GSH is consumed by oxidants and immediately regenerated through the redox buffering system by glutathione reductase and NADPH (
1). Stem cells defective in GSH regeneration exhibit reduced functional activity and therapeutic effects (
3).
GSH is synthesized by glutamate-cysteine ligase and glutathione synthetase, which are expressed by transcri-ption factors (such as NRF2) under oxidative stress. GSH is synthesized in the cytosol and distributed in subcellular organelles such as nucleus, mitochondria, endoplasmic reticulum, and Golgi apparatus where it functions differentially in each organelle. For example, mitochondrial GSH (mGSH) plays crucial roles in cell death and human diseases (
6,
7). Therefore, the comprehensive and organelle-specific GSH assay methods need to be developed for monitoring GSH levels in stem cells.
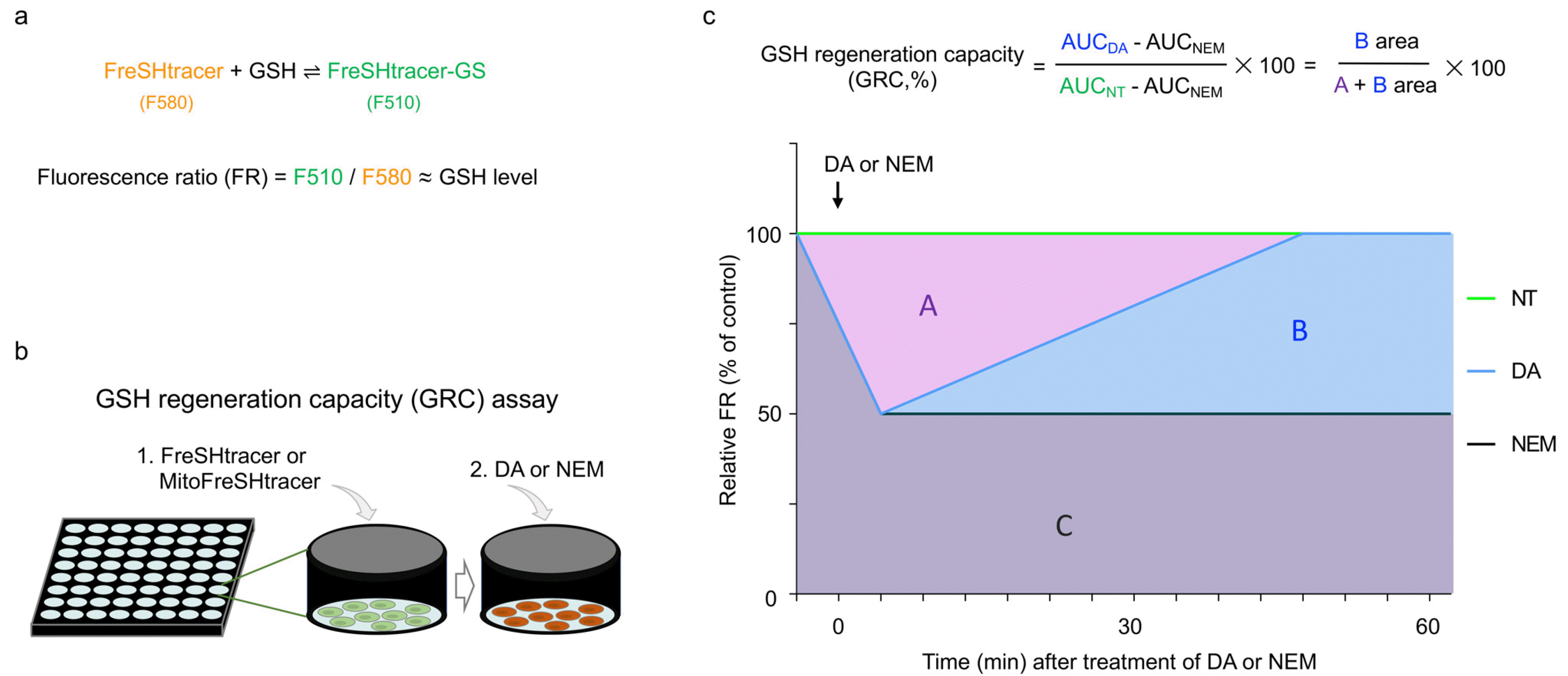
For the first time, we have developed Fluorescent real-time thiol tracer (FreSHtracer) for real-time monitoring of GSH in living cells (
3,
8,
9). FreSHtracer is a ratiometric probe that reversibly and rapidly reacts with GSH (
Fig. 1a). Since it is cellular membrane permeable, FreSH-tracer-based GSH monitoring of living cells is feasible (
Fig. 1b). FreSHtracer is modifiable and can be used for monitoring of organelle-specific GSH such as MitoFreSH-tracer for mGSH (
3,
9). Thus, FreSHtracer-based monitoring is an ideal GSH-monitoring system suitable for living cells.
In this report, we describe a method for quantifying the GSH regeneration capacity (GRC) in cultured stem cells using FreSHtracer and MitoFreSHtracer. This protocol has been devised to measure GRC levels in human umbilical cord-derived mesenchymal stem cells (hUC-MSCs) and human bone marrow-derived mesenchymal stem cells (hBM-MSCs). Furthermore, we believe that with minor modifications detailed here, this protocol can be applied to all adherent cultured mammalian cells.
Materials and Methods
Reagents
FreSHtracer (cat. No. F-402, Cell2in, Korea).
MitoFreSHtracer (cat. No. M-377, Cell2in).
Cells of interest: hUC-MSCs (10) and hBM-MSCs (cat. No. PT-2501, Lonza). CAUTION: These cells need to be regularly monitored to ensure that they are free of Mycoplasma infection.
Minimum Essential Medium (MEM) Alpha (cat. No. 12561056, Gibco).
Fetal bovine serum (FBS; cat. No. 16000044, Gibco).
Penicillin/streptomycin, 100× (cat. No. 15140122, Gibco).
Mesenchymal stem cell basal medium (MSCBM; cat. No. PT-3238, Lonza).
MSCGM SingleQuats (cat. No. PT-4105, Lonza).
Hanks’ Balanced Salt Solution (HBSS; cat. No. LB 003-02, WELGENE).
Dimethyl sulfoxide (DMSO; cat. No. 1.02931.1000, Merk Millipore).
Dulbecco’s Phosphate buffered saline (PBS) 1× sterile solution (DPBS, pH 7.4; cat. No. LB 001-02, WEL-GENE).
HEPES (cat No. 25-060-Cl, CORNING).
TrypLETM Express Enzyme, 1×, no phenol red (cat. No. 12604-013, Gibco).
Trypan blue (cat. No. T8154, Sigma Aldrich).
Diamide (DA; cat No. D3648, Sigma-Aldrich).
Water, Ultra Pure (cat. No. W2006, Biosesang).
Happy cell (incubator water; cat. No. BHC002, Biomax).
N-Ethylmaleimide (NEM; cat No. E1271, Sigma-Aldrich).
Equipment
High-content screening (HCS) microscopy (Operetta, PerkinElmer) was performed using the Harmony High-Content Imaging and Analysis Software (PerkinElmer).
Centrifuge (Eppendorf, cat. No. 5810R).
Centrifuge tube (15 ml; cat. No. 50015, SPL).
Centrifuge tube (50 ml; cat. No. 50050, SPL).
Cell culture dish (100 mm; cat. No. 430167, Corning).
A Pipet-Aid XP (cat. No. 4-000-201-E, Drummond Scientific Co., Ltd.).
Serological pipette (cat. No. 91005, 91010, 91025; SPL).
Micropipettes (cat. No. 4641060, 4641080, 4641100; Thermo Scientific).
Multi Channel Pipettors, 10∼100 μl (cat. No. 312200 0043, Eppendorf).
Micropipette tips (cat. No. 15730, 10040, Sorenson).
Reservoir (cat. No. 23050, SPL).
0.2 μm syringe filter (cat. No. STI16534-K, Sartorius).
96-well flat bottom black polystyrene cell culture plate (cat. No. 655090, Greiner Bio-One).
Hemocytometer (cat. No. 0610039094, Marienfeld).
Aspiration pump (cat. No. JSBS-5000, JRS).
Fluorescence-activated cell sorting (FACS) tubes (5 ml Polystyrene Round-Bottom Tube, 12×75 mm; cat. No. 352054, FALCON).
FACS filter tubes (5 ml, polystyrene round-bottomed tube with cell-strainer cap; cat. No. 352235, FALCON).
Flow cytometer (BD FACS LSRFortessa).
Reagent setup
FreSHtracer and MitoFreSHtracer: The stock solutions were prepared as described previously (
10).
Culture medium: hUC-MSCs were cultured in MEM Alpha supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. The hBM-MSCs were incu-bated in MSCBM supplemented with MSCGM SingleQuats. The medium need to be stored at 4℃ until expiration.
FACS solution: For flow cytometry, the cells were suspended and analyzed in DPBS containing 2% FBS.
Equipment setup
HCS microscopy system for measuring GRC: The confocal imaging system consists of Operetta High-Content Imaging System with ×200 or ×400 magnifications. Fluo-rescence images were detected at emission (Em) wavelengths in the range of 500∼540 and 560∼630 nm and excitation (Ex) wavelengths in the range of 410∼430 and 490∼510 nm. These measurements were used to obtain the fluorescence ratio (F510/F580, FR) for FreSHtracer or MitoFreSHtracer (
4,
9). The HCS instrument was prewarmed for at least 30 min.
Software for GRC analysis: The fluorescence intensities were determined using Harmony High-Content Imaging and Analysis Software 4.1. The FR values per well were calculated by dividing the F510 (Ex410∼430, Em460∼540) by the F580 (Ex490∼510, Em560∼630) inten-sities. The area under the curve (AUC) values over time were determined using GraphPad Prism 8.0 software (Gra-phPad Software). The GRC was calculated using the formula shown in
Fig. 1c.
Flow cytometry: A flow cytometry system was used to obtain the FR values, as previously described (
9). Briefly, the cells were loaded with 5 μM of FreSHtracer or Mito-FreSHtracer and subjected to analysis using the two channels of F510 (Ex405, Em475∼575) and F580 (Ex488, Em550∼600 or Ex561, Em567∼597) and gating with forward scatter area and side scatter area/height/width. The mean and heterogeneity of the FR values were evaluated by applying the geometric mean and robust coefficient of variation, respectively, using the FlowJo program.
Statistical analysis
The data were analyzed using a two-sided t-test for single pairwise comparison and a nonparametric Mann-Whitney U test or analysis of variance (ANOVA) (one- or two-way) with a Bonferroni post hoc test for multiple com-parisons. All statistical analyses were performed using GraphPad Prism software (version 8.0; GraphPad Software, San Diego, CA, USA).
Procedure
GRC measurement in living stem cells using HCS microscopy
1│ The cells of interest were plated on the 96-well flat bottom black polystyrene cell culture plates. ▲ CRITICAL STEP: Adjusting the cell seeding number is critical because confluency regulates the intracellular GSH levels (3). In the case of the hUC-MSCs and hBM-MSCs, they were seeded onto the plates at a density of 3∼4×103 cells/well in 100 μl culture medium in each well for 1 day.
2│ FreSHtracer (option A) or MitoFreSHtracer (option B) was added onto the cells, as previously described (10). Briefly, (A) the cells are incubated with 5 μM of FreSHtracer for 1.5 h at 37℃ and 5% CO2. Then, the GRC was measured. (B) The cells were incubated with 10 μM MitoFre-SHtracer for 1.5 h at 37℃ and 5% CO2. The cells were washed twice with HBSS, and 2 ml of a new medium without MitoFreSHtracer was added.
3│ An HBSS-based solution containing 10 mM HEPES was prepared for performing HCS microscopy. (A) (i) A dilution solution containing 5 μM FreSH-tracer in the HBSS-based solution was used. (ii) Oxidant solutions contained 0.1 mM NEM or various concentrations of DA in the dilution solution were used.(B) (i) The HBSS-based solution was used as the washing and dilution solutions. (ii) Either a 0.1 mM NEM or various concentrations of DA in the HBSS-based solution were used as oxidant solutions. ▲ CRITICAL STEP: As NEM vapor can affect the quality of the other solutions used for cellular GSH measurements, the NEM working solution was prepared separately from the other solutions. •WARNING: As continuous inhalation of NEM vapor may cause corrosive injuries to the upper airways and lungs, caution must be exercised when handling the NEM solution.
4│ All solutions were poured into the reservoirs or new 96-well plates to enable easy handling of the multi Channel pipettors.
5│ The F510 and F580 intensities in each well were examined using HCS microscopy. ▲ CRITICAL STEP: The fluorescence intensities that generally represent cell count/well should not be differ among the wells.
6│ The culture media were suctioned out of the wells using vacuum generated by an aspiration pump. (B) The cells were washed twice with HBSS.
7│ Next, 40 μl of oxidant solution/well was added immediately using multi Channel pipettors. The cells were incubated for 10 min at 37℃.
8│ Following incubation, 160 μl of dilution solution was added per well.
9│ The F510 and F580 intensities/well were measured using HCS microscopy every 3 min for 1 h.
Timing
Step 1, cell plating: 1 day
Step 2, loading probes into cells: 1.5 h
Steps 3∼9, measuring GRC in living cells: <2 h
Results
Establishment of GRC assay
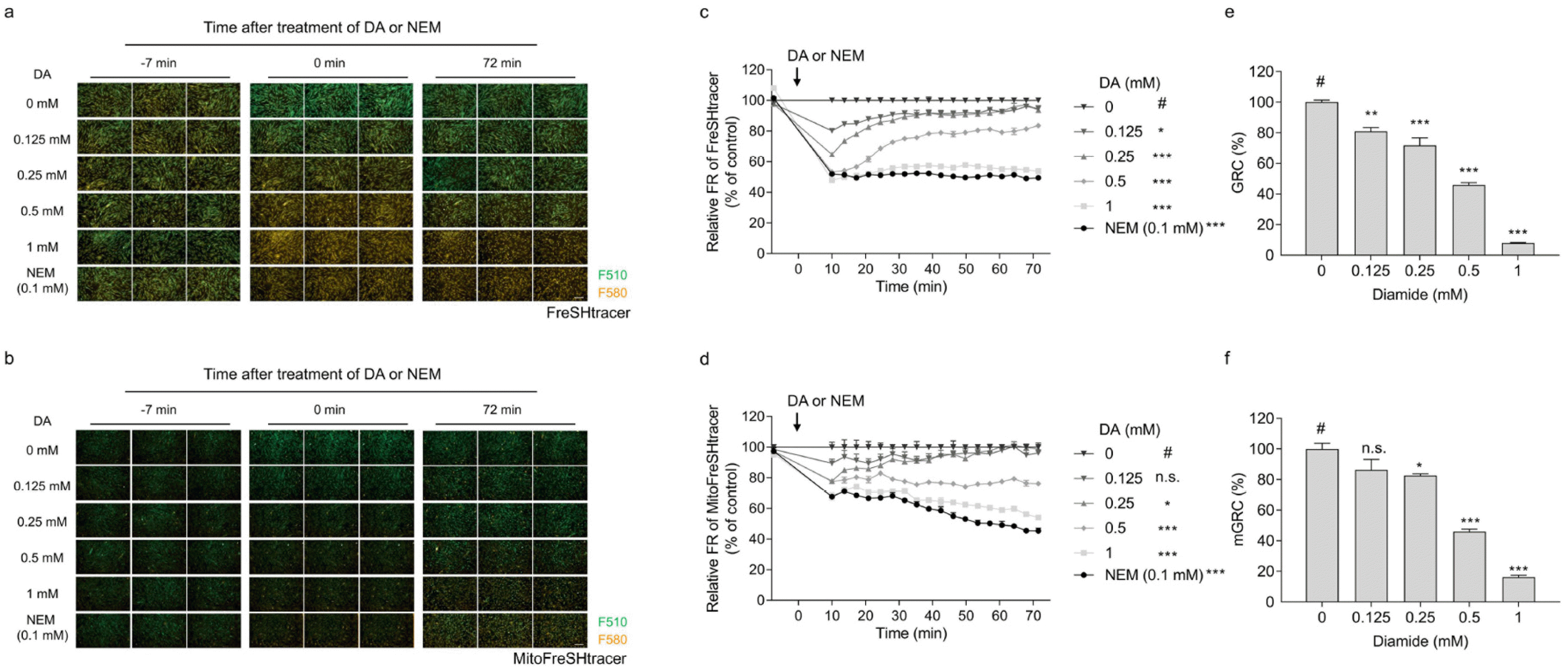
hUC-MSCs were used for establishing the GRC assay. FreSHtracer-stained cells were treated with the indicated concentrations of DA or NEM (0.1 mM), and the fluorescence intensities of the cells were observed using confocal microscopy (
Fig. 2a and
2b). As NEM is irreversibly incorporated into thiols in cells, the NEM-treated group was used to estimate the cellular basal FR values. The FR values were plotted over time for approximately 1 h (
Fig. 2c and
2d). The GRC was determined by dividing the difference of the AUCs of the non-treated group and 0.1 mM NEM-treated group by the difference of the AUCs of the DA-treated group and 0.1 mM NEM-treated group (
Fig. 1c). The GRC levels in hUC-MSCs decreased in a dose-dependent manner following treatment with DA (
Fig. 2e and
2f), implying that the GRC assay is applicable to hUC-MSCs.
Analyzing GSH levels of whole cellular and mitochondrial areas using flow cytometry
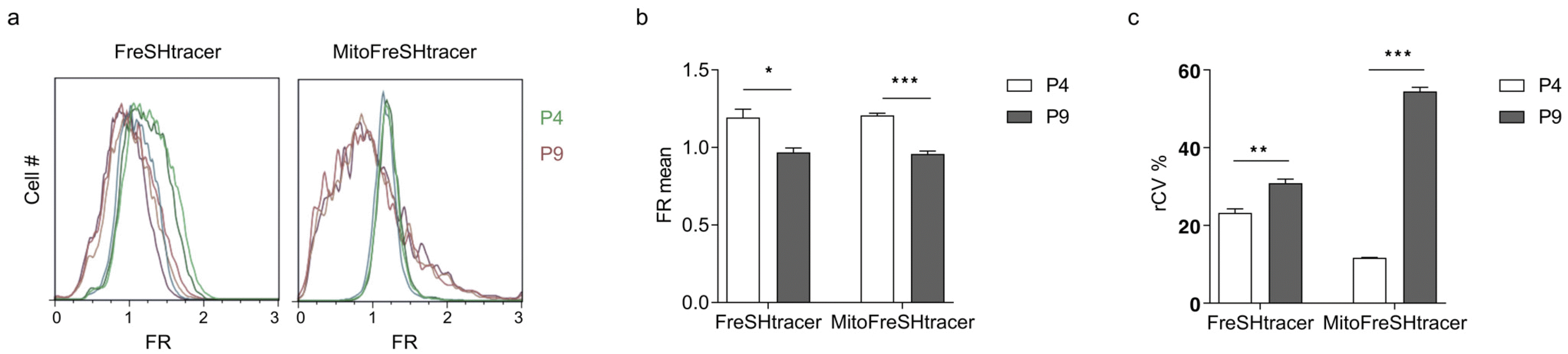
Previously, we showed that GSH levels decreased and GSH heterogeneity increased in hBM-MSCs as the passage number increased (
3). We used this finding to analyze the mean and heterogeneity of the GSH levels pertaining to the whole-cell and mitochondria in hBM-MSCs with different passage numbers (P4 and P9) using flow cytometry. P4 hBM-MSCs exhibited significantly higher FR mean values and lower FR robust coefficient of variation (rCV) values than those at P9 (
Fig. 3). Interestingly, the rCV levels for MitoFreSHtracer were approximately 4.7 times different between the P4 and P9 cells. This indicates that the rCV values of MitoFreSHtracer were more sensitive to the passage-dependent conditions of hBM-MSCs than those of FreSHtracer, which stains GSH in the entire cell.
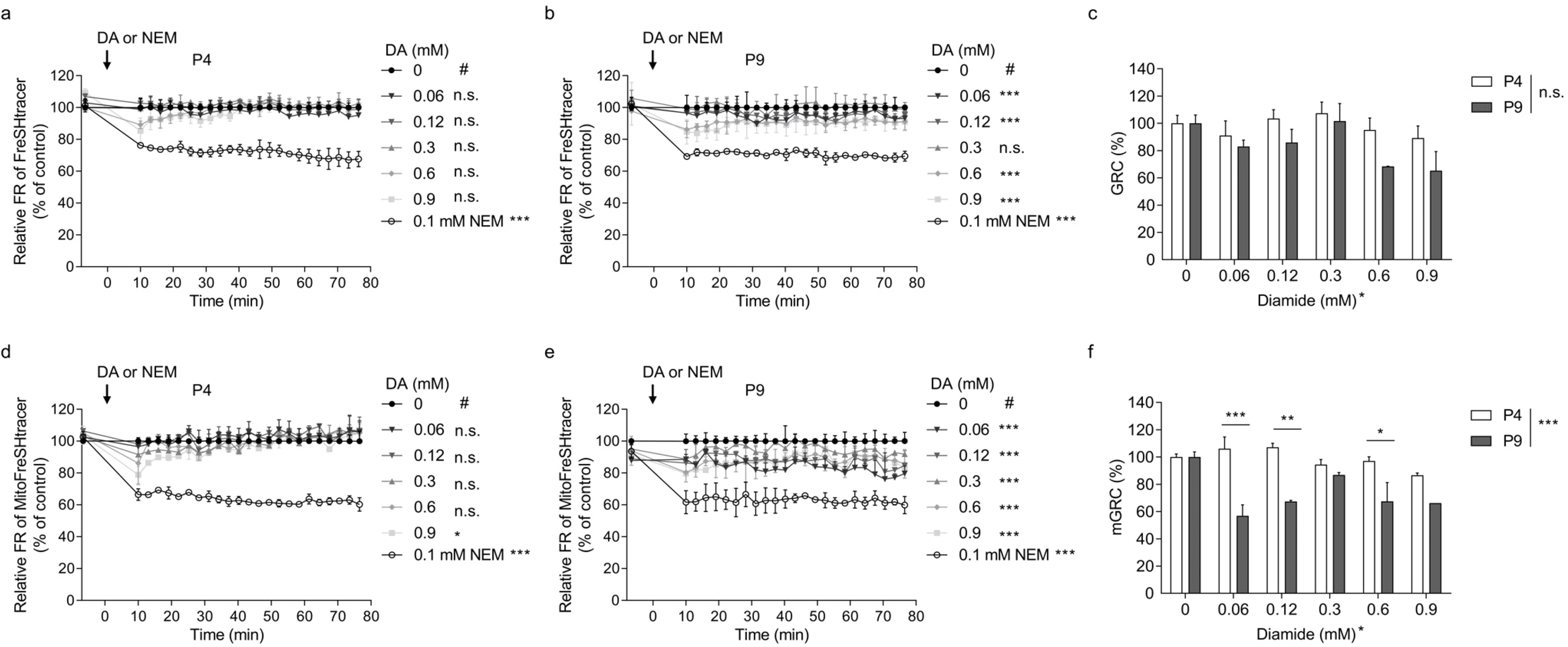
Measuring GRC of whole cellular and mitochondrial areas using HCS microscopy
The P4 and P9 hBM-MSCs were used for the GRC assay. The GRC levels for the whole cellular and mitochondrial areas were analyzed using FreSHtracer and MitoFreSHtracer, respectively. Whereas P4 cells showed no significant kinetic changes in the FR of the whole cell area, P9 cells exhibited significant changes at all DA concentrations except at 0.3 mM (
Fig. 4a and
4b). The GRC values were calculated based on the FR kinetics. However, the values were not significantly different between P4 and P9 cells (
Fig. 4c).
Investigation of the mitochondrial GRC in hBM-MSCs showed that the FR kinetics of the P4 cells were significantly altered only in the cells treated with 0.9 mM DA (
Fig. 4d), whereas those of P9 cells were significantly altered at all DA concentrations (
Fig. 4e). Furthermore, the GRC values were significantly different between both cells at all treatments except for the 0.3 mM DA-treated groups (
Fig. 4f).
Discussion
In this study, we developed a method to quantify the GRC in mesenchymal stem cells using a FreSHtracer-based HCS microscopy system. The GRC assay is typically performed within 4 h of cell seeding. We could significantly distinguish the P4 and P9 hBM-MSCs by measuring the mitochondrial GRC (mGRC), but not the whole-cell GRC. Thus, this protocol is simple, rapid, and relia-ble. It is a promising tool for enhancing the efficacy of therapeutic stem cells.
Previously, we had reported a GRC assay based on the GSH recovery slope and initial GSH levels using FreSH-tracer (
4). We have upgraded the current methodology by including the area under the curve analysis and MitoFre-SHtracer probe to the protocol to obtain a more comprehensive and reliable GRC quantification data compared with that obtained using the previous method.
DA is a thiol-specific oxidant that decreased the GRC levels in hBM-MSCs under most conditions. Interestingly, the decrease in GRC caused by DA treatment was not always concentration-dependent. For example, the mGRC of the P9 cells treated with 0.06 mM DA, which was the lowest treatment concentration, showed the most effective decrease in the GRC value (
Fig. 4f). Moreover, the mGRC values of the P9 cells gradually increased in a dose dependent manner as the DA-treatment concentration increased from 0.06 to 0.3 mM. This can probably be attributed to the threshold at which the expression of genes related to the cellular redox buffering system is activated by the KEAP1-NRF2 signaling pathway; however, further study is required to confirm this hypothesis.
GSH is synthesized in the cytosol and transferred to other organelles, including the nucleus, mitochondria, endoplasmic reticulum, and Golgi complex. Given that the mitochondria generate reactive oxygen species during oxidative phosphorylation to generate ATP, mGSH is critically involved in reducing lipid peroxidation and regulating cell death (
6). The present study showed that the mGRC and heterogeneity of mGSH were highly sensitive to the passage-dependent conditions of MSCs (
Fig. 3c and
4f), implying that the mGSH redox buffering capacity might be crucially related to stem cell quality. Future studies are necessary to investigate whether this GRC assay can identify other critical culture factors that impact stem cell efficacy such as hypoxia and donor age. Further studies need to be performed to determine if this GRC assay may be applied to other stem cells and ascertain the sensitivity of other organelles to the stem cell quality. Thus, the GRC may be a good index for evaluating stem cell quality.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download