Abstract
Background and Objectives
Human induced pluripotent stem cell (hiPSC)-derived cardiomyocyte (CM) hold great promise as a cellular source of CM for cardiac function restoration in ischemic heart disease. However, the use of animal-derived xenogeneic substances during the biomanufacturing of hiPSC-CM can induce inadvertent immune responses or chronic inflammation, followed by tumorigenicity. In this study, we aimed to reveal the effects of xenogeneic substances on the functional properties and potential immunogenicity of hiPSC-CM during differentiation, demonstrating the quality and safety of hiPSC-based cell therapy.
Methods and Results
We successfully generated hiPSC-CM in the presence and absence of xenogeneic substances (xeno-containing (XC) and xeno-free (XF) conditions, respectively), and compared their characteristics, including the contractile functions and glycan profiles. Compared to XC-hiPSC-CM, XF-hiPSC-CM showed early onset of myocyte contractile beating and maturation, with a high expression of cardiac lineage-specific genes (ACTC1, TNNT2, and RYR2) by using MEA and RT-qPCR. We quantified N-glycolylneuraminic acid (Neu5Gc), a xenogeneic sialic acid, in hiPSC-CM using an indirect enzyme-linked immunosorbent assay and liquid chromatography-multiple reaction monitoring-mass spectrometry. Neu5Gc was incorporated into the glycans of hiPSC-CM during xeno-containing differentiation, whereas it was barely detected in XF-hiPSC-CM.
Conclusions
To the best of our knowledge, this is the first study to show that the electrophysiological function and glycan profiles of hiPSC-CM can be affected by the presence of xenogeneic substances during their differentiation and maturation. To ensure quality control and safety in hiPSC-based cell therapy, xenogeneic substances should be excluded from the biomanufacturing process.
Heart disease is a leading cause of morbidity and mortality globally (1). Treatment with drugs and surgical intervention does not affect the causal factor of cardiac function impairment—a major loss of cardiomyocyte (CM) after ischemic injury. In recent times, human pluripotent stem cell (hPSC)-based therapy has emerged as a promising approach for the treatment of heart disease (2-4). Although hPSC-derived CM (hPSC-CM) has been considered an attractive cell source for cardiac remuscularization (5), the use of animal-derived xenogeneic substances, such as fetal bovine serum (FBS), porcine trypsin, and MatrigelTM, in the differentiation culture process could hamper the clinical outcomes. Reportedly, the use of xenogeneic substances in the maintenance and differentiation of stem cells can trigger unexpected immune responses followed by inflammation (6, 7). To overcome this, the use of chemically defined and/or xeno-free (XF) culture systems have been suggested for hPSC colony expansion and differentiation (8, 9). However, it is necessary to verify the characteristics and safety issues associated with xenogeneic substances when hPSC-CM is used for cell therapy, especially in the differentiation and biomanufacturing processes.
N-glycolylneuraminic acid (Neu5Gc), a non-human sialic acid, has been identified as an immunogenic xeno-antigen because it is not synthesized in human cells (10, 11). In mammals, the terminal sialic acid N-acetylneuraminic acid (Neu5Ac) and its derivative Neu5Gc are the two most common forms of a 9-carbon monosaccharide (12). Owing to loss-of-function mutations in the gene encoding cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMAH), an enzyme that converts Neu5Ac to Neu5Gc, human cells cannot synthesize Neu5Gc, which increases the abundance of its precursor Neu5Ac (11). Exogenous Neu5Gc derived from animal-derived food products, such as red meat and dairy milk, can be incorporated into newly synthesized glycans in human cells, which can induce the production of anti-Neu5Gc antibodies in circulation (13-15). The anti-Neu5Gc immune response promotes cancer-related inflammation, leading to cancer progression (14, 16-18). A study demonstrated that Neu5Gc-containing biotherapeutics may induce immune response-driven chronic inflam-mation, known as “xenosialitis” (6). Therefore, it is important to eliminate xenogeneic substances during the manufacturing of biotherapeutics, especially during cell culture and/or differentiation in cell-based therapies.
In this study, we generated human induced pluripotent stem cell (hiPSC)-derived CM (hiPSC-CM) in the presence or absence of xenogeneic substances using modified methods of cell differentiation (19). Additionally, based on concerns regarding the safety of hiPSC-CM, we demonstrated that xenogeneic Neu5Gc can be incorporated into hiPSC-CM from media containing xenogeneic substances (xeno-containing, XC) during differentiation, which indicated the importance of excluding xenogeneic substances from the biomanufacturing process of hiPSC-CM.
hiPSC (FSiPS1; obtained from the National Stem Cell Bank, KNIH, Korea) were cultured in E8 medium (Gibco, Carlsbad, CA, USA) under XF conditions or in mTeSRTM1 (STEMCELL Technologies, Vancouver, Canada) under XC conditions at 37℃ in the presence of 5% CO2. As a coating material for the 6-well plate, Vitronectin (VTN-N) Recombinant Human Protein (Gibco) was used in the XF culture and MatrigelTM hESC-qualified (Corning, Bedford, MA, USA) was used in the XC culture. The culture media were replaced daily, and the cells were subcultured (at a 1:7 ratio) using a non-enzymatic cell dissociation method with Versene solution (Gibco) every 5 days.
hiPSC-CM was generated by inducing the differentiation of hiPSC into cardiac-lineage cells under XF or XC conditions (Fig. 1). For XC culture, the previously reported GiAB protocol (MM.1) (19) was used, with some modifications by adding matrix MatrigelTM at day 0 and day 1 of differentiation. Briefly, 5×105 hiPSCs/well were seeded on a MatrigelTM-coated 12-well plate and treated with mTeSRTM1 supplemented with the GSK3 inhibitor CHIR99021 (1 μM) for 4 days. To induce the differentiation of hiPSC into mesodermal cells and cardiac-lineage cells, we used RPMI-based differentiation media, which contain various components, such as activin A, basic fibroblast growth factor (bFGF), knock-out serum replacement (KO-SR), B-27 supplements-insulin, and BMP4, in different combinations, depending on the differentiation stage. For XF culture, according to the manufacturer’s instructions, hiPSC were differentiated into CM using the Gibco PSC Cardiomyocyte Differentiation Kit (Gibco). Briefly, human iPSCs were singularized using ACCUTASETM, and 1×105 cells/well were seeded on a VTN-N-coated 12-well plate in E8 medium. The E8 medium was replaced daily for 4 days to allow sufficient proliferation of hiPSC, and then the following media were applied sequentially: cardiomyocyte differentiation medium A, cardiomyocyte differentiation medium B, and cardiomyocyte mainte-nance medium. Ten days after differentiation, the beating of the CM was observed. All cultures were maintained at 37℃ in the presence of 5% CO2.
Total RNA was extracted from hiPSC-CM under XF or XC conditions at 0, 2, and 4 weeks after differentiation using the RNeasy Plus Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s recommen-dations. After the quantification of RNA, complementary DNA (cDNA) was synthesized using 1 μg of RNA and an iScriptTM cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s ins-tructions. RT-qPCR was performed using a cDNA template and the QuantiTect SYBR Green PCR Kit (QIAGEN) in a real-time PCR system (7900HT Fast Real-Time PCR System, Applied Biosystems, Waltham, MA, USA). The sequences of primers used for the target genes are listed in Supplementary Table S1. The relative expression of genes was calculated using the 2–ΔΔCtmethod with ExpressionSuite Software Version 1.2 (Thermo Fisher Scientific, Waltham, MA, USA). The expression values were normalized against those of the 18S rRNA gene as a housekeeping gene.
hiPSC-CM were fixed in 4% paraformaldehyde (Sigma, St. Louis, MO, USA) for 10 min and permeabilized with 0.1% Triton X-100 (Sigma) in PBS for 10 min. After the cells were treated with an immunofluorescence blocking buffer (Cell Signaling Technology, Danvers, MA, USA), they were treated overnight at 4℃ with the following primary antibodies: MLC-2v (1:200, Thermo Fisher Scientific, 10906-1-AP), Troponin T (1:200, ab8295, Abcam, Cam-bridge, UK), α-SMA (1:200, Cell Signaling Technology, #19245s), and α-actinin (1:100, MERCK, A7811). All primary antibodies were diluted using an immunofluo-rescence antibody dilution buffer (Cell Signaling Technology). After washing with a 1X washing buffer (Cell Signaling Technology), the cells were incubated for 1 h at room temperature with goat anti-rabbit IgG (H+L)-Alexa Fluor 555 (1:250, Thermo Fisher Scientific, A21428) and goat anti-mouse IgG (H+L)-Alexa Fluor 488 (1:250, Thermo Fisher Scientific, A11001) as fluorescence-conjugated secondary antibodies. The nuclei were stained with DAPI (NucBlueTM Fixed Cell Stain ReadyProbesTM Reagent, Thermo Fisher Scientific, R37606). Images of the cells were acquired using a fluorescence microscope (Axio Observer 7; Carl Zeiss).
This assay was performed as previously described (20). In brief, to attach hiPSC-CM on the assay plate, 12-well MEA plates (Axion Biosystems, Atlanta, GA, USA) containing electrodes were coated for 30 min with 50 μg/ml fibronectin in DPBS (7 μl/well). hiPSC-CM were singularized with the STEMdiffTM Cardiomyocyte Dissociation Kit (STEMCELL Technologies, Vancouver, Canada), and 4×106 cells were seeded in plating media suitable for XC- or XF-hiPSC-CM maintenance and transferred to the MEA plate. Field potentials (FPs) were recorded for spontaneously beating hiPSC-CM. During the recording period, the MEA plate was placed at 37℃ in a sterile environment using the Maestro MEA system (Axion Biosystems). After the FP waveforms were stabilized, the signals were recorded for 5 min and the last 1 min were analyzed. Fridericia’s formula (FPDcF), where FPDcF=field potential duration (FPD)/beat period0.33, was used to correct the dependence of the FPD on the beating rate.
According to the manufacturer’s recommendations, Neu5Gc was detected using an ELISA with an Anti-Neu5Gc antibody kit (BioLegend, San Diego, CA, USA; free of Neu5Gc). Briefly, serum (10 μl/well), conditioned medium (10 μl/well), and cell lysate (500 μg/well) samples collected under XC or XF conditions were placed on a high-binding 96-well ELISA plate (Corning). After the sodium carbonate-bicarbonate immobilization buffer (50 mM, pH 9.5, Invitrogen, Carlsbad, CA, USA) was added to each well, the samples were incubated for 16∼18 h at 4℃. The wells were blocked for 1 h at room temperature with the Neu5Gc Assay Blocking Solution and incubated for 2 h with a purified anti-Neu5Gc antibody (primary antibody) and chicken IgY isotype control diluted at a ratio of 1:5,000 in PBS TweenTM 20 buffer (PBST) (Pierce Biotechnology, Rockford, IL, USA). The plates were washed three times with PBST and subsequently incubated for 1 h at room temperature with goat anti-chicken IgY H&L (HRP) (Abcam) diluted at a 1:5,000 ratio in PBS. After washing three times with PBST, the wells were developed with a substrate reagent pack and the stop solution 2N sulfuric acid (R&D Systems, Minneapolis, MN, USA). Absorbance was measured at 490 nm using the Spectra-Max 190 with SoftMax Pro 6.3 software (Molecular Devices).
hiPSC-CM lysates were prepared at 0, 2, 4, and 6 weeks of differentiation in the presence or absence of xenogeneic substances. To quantify sialic acids (Neu5Gc and Neu5Ac) in hiPSC-CM, cell membranes were isolated and purified using PGC-SPE according to the method described in a previous report (21). The enriched sialic acids were analyzed using an Agilent 1290 Infinity LC system coupled with an Agilent 6495 triple quadrupole mass spectrometer. An Agilent AdvanceBio MS Spent Media column was used for LC separation with a binary gradient composed of solvent A (100% water and 10 mM ammonium formate (v/v) and solvent B (90% ACN with 10 mM ammonium formate (v/v) at a flow rate of 0.3 ml/min. N-linked glycans were released enzymatically from the prepared cell membranes by treating with PNGaseF and further enriched by solid-phase extraction (SPE), as previously described (22). Purified N-glycans were comprehensively analyzed using an Agilent 1260 Infinity LC and Agilent 6530 Q-TOF MS system. The abundance of each glycan was normalized to the total abundance of all observed glycans (23).
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA). The standard unpaired Student’s t-test was used for comparisons between two groups. Analysis of variance followed by Tukey’s multiple comparisons test was used to compare more than two groups. RT-qPCR and cell biological assays were performed at least in triplicate with three to five independent experiments. Data are presented in terms of means±SEM. Statistical significance was set at p<0.05.
To compare the characteristics of hiPSC-CM depending on xenogeneic substances, we differentiated hiPSC into CM in the presence and absence of xenogeneic substances, such as MatrigelTM, FBS, porcine trypsin, and bovine serum albumin. XC- and XF-hiPSC-CM were successfully generated using our modified differentiation method (Fig. 1). After the hiPSCs were expanded for 4 days on MatrigelTM-coated plates (XC) or VTN-N-coated plates (XF), mesoderm induction occurred within 2 days after the media were changed under both XC and XF conditions. The addition of MatrigelTM to the culture media enhanced cell attachment and mesoderm induction (XC conditions). However, differentiation into cardiac-progenitor cells occurred earlier upon VTN-N coating (XF conditions) (day 4) than under XC conditions (day 7). The expression of ISL1, a mesoderm marker, peaked at 2 weeks in both XC- and XF-hiPSC (Supplementary Fig. S1). The maturation of early-stage CM with contractile beating activity occurred more rapidly in the XF condition than in the XC condition. The contractile beating of hiPSC-CM with multiple contractile points was observed from 2 weeks after differentiation under both culture conditions, and synch-ronized beatings were observed at 4 weeks (Fig. 2A; Supplementary Video S1). After 4 weeks of differentiation, hiPSC-CM gathered and detached from the base of the plate, gradually forming a reticulated cardiac sheet. We continuously induced CM’s differentiation for further maturation into late-stage CM until 6 weeks. From these results, we demonstrated that the generation of XF-hiPSC-CMs with synergistic beating activities is robust and has even more rapid progress in differentiation from hiPSCs compared to XC-hiPSC-CM.
To evaluate the maturation status of XC- and XF-hiPSC-CM, we first measured cardiac-lineage mRNA expression using reverse transcription-quantitative PCR (RT-qPCR). The expression of myocardial sarcomeric protein-coding genes (MYH7, MYL2, ACTC1, and TNNT2) increased at 2 and 4 weeks after differentiation into cardiac-lineage cells under both conditions (Fig. 2B). At two weeks of differentiation, the fold changes in the expression of cardiac lineage-specific genes (MYH7, ACTC1, and TNNT2) increased in XF-hiPSC-CM compared to that in XC-hiPSC-CM (Fig. 2B, left panel).
The mRNA expression of cardiac ion channel-related genes, such as hyperpolarization-activated cyclic nucleotide-gated potassium channel 4 (HCN4) and ryanodine receptor 2 (RYR2), was significantly increased in both XC- and XF-hiPSC-CM compared with hiPSC. At two weeks of differentiation, RYR2 expression was significantly higher in XF-hiPSC-CM than in XC-hiPSC-CM (p<0.05, n=4) (Fig. 2B, right panel). In contrast, the expression of POU5F1, a marker of undifferentiated iPSC, was downregulated in both XC- and XF-hiPSC-CM after 2 to 4 weeks of differentiation. At 2 weeks, the expression of ISL1, a mesoderm marker, was higher in hiPSC-CM than in hiPSC; however, it was reduced by 4 weeks of differentiation under both XC and XF conditions (Supplementary Fig. S1). Next, we examined cardiac lineage-specific protein expression in hiPSC-CM. Immunocytochemistry confirmed the expression of cardiac sarcomere components and contractile cytoskeletal proteins, including ventricular myosin light chain (MLC-2v), alpha-smooth muscle actin (α-SMA), α-actinin, and cardiac troponin T (cTNT), in hiPSC-CM (Fig. 2C). These results demonstrated that the XF-hiPSC-CM shows characteristics of CMs with high expression of cardiac-specific genes and proteins, therefore suggesting differentiation into mature CMs at the molecular levels.
Prolonged culture after the induction of cardiac differentiation from stem cells can improve cardiac cell maturity with respect to morphology and electrophysiology (24-26). To compare the contractile functions of XF- and XC-hiPSC-CM, their electrophysiological properties were analyzed by using an in vitro multi-electrode array (MEA) system at 2 and 4 weeks of differentiation (Fig. 3). The representative field potential signals recorded from approximately 4 weeks of XC- and XF-hiPSC-CM were shown in Supplementary Fig. S2. In XC-hiPSC-CM, all analyzed field potential parameters increased significantly at 4 weeks from that at 2 weeks (Fig. 3A). The BPMs increased significantly from 33.3±2.38 bpm at 2 weeks to 52.77±4.08 bpm at 4 weeks (n=6, **p=0.0015). The FPA increased considerably, from 0.4±0.02 mV at 2 weeks to 1.56±0.15 mV at 4 weeks (n=6, ***p<0.0001). The FPDcF also increased from 319.8±11.27 ms at 2 weeks to 502.6±36.99 ms at 4 weeks (n=6, ***p=0.0008). In XF-hiPSC-CM, the beats per min (BPMs) increased significantly, from 43.2±1.52 bpm at 2 weeks to 52.4±2.6 bpm at 4 weeks (number of wells at each time point (n)=6, *p=0.0129). The field potential amplitude (FPA) also increased significantly, from 1.04±0.1 mV at 2 weeks to 2.01±0.17 mV at 4 weeks (n=6, ***p=0.0005). The field potential duration (FPDcF) did not change signifi-cantly, but showed an increasing trend, from 434.2±21.5 ms at 2 weeks to 466.6±18.07 ms at 4 weeks (Fig. 3B).
The electrophysiological parameters of hiPSC-CM were also compared to those of the commercial cardiomyocyte NexelTM CardiosightⓇ-S after 4 weeks of differentiation (Fig. 3C). Even though the FPA of NexelTM CardiosightⓇ-S was 4.28±0.4 mV, which was significantly higher than those of XF- or XC-hiPSC-CM (n=4, ***p<0.0001), the BPM and FPDcF of XF- or XC-hiPSC-CM did not differ significantly from those of NexelTM CardiosightⓇ-S. Con-versely, undifferentiated hiPSC cultured in XC or XF medium did not show electric excitability in the MEA system (data not shown). From the results of MEA analysis, we demonstrated that the XF-hiPSC-CMs also have mature electrophysiological characteristics of CMs, showing higher contractile activities compared to XC-hiPSC-CMs.
Based on the results of the characteristic analysis, both XC- and XF-hiPSC-CM exhibited characteristics of mature CM, with contractile function induced at 4 to 6 weeks of differentiation. Owing to safety concerns regarding the induction of a xenogeneic immune response, we investigated whether Neu5Gc, a xenogeneic sialic acid in humans, can be incorporated into terminal glycans in XC-hiPSC-CM. During the maintenance and cardiac-lineage differentiation of hiPSC into CM, we examined the Neu5Gc content in complete medium, conditioned medium, hiPSC lysates, and hiPSC-CM lysates using an indirect enzyme-linked immunosorbent assay (ELISA). For comparison of the XC/XF samples, FBS was used as the positive control and internal standard for Neu5Gc. As shown in Fig. 4A, Neu5Gc was detected in all XC media. The Neu5Gc content was significantly higher in the CM differentiation media (XCMdiff A and XCMdiff B) than in the hiPSC-culture media (mTeSR1) or cardiomyocyte maintenance media (XCM). In contrast, Neu5Gc was not detected in the XF media, such as the Essential 8 (E8) medium, CMdiff A, CMdiff B, and CM_main (Fig. 4A). Among the XC media obtained from the cell culture and differentiation experiments, the hiPSC-CM differentiation media showed a significantly higher Neu5Gc content (at 2 to 4 weeks vs. 0 weeks, n=3, *p<0.05) than the hiPSC culture media. However, Neu5Gc was not detected at any time point in the XF medium. Interestingly, we found that the incorporation of Neu5Gc into XC-hiPSC-CM was maintained during weeks 2 to 6 of differentiation (Fig. 4B). Using ELISA, we detected Neu5Gc, a non-human sialic acid form, in XC-hiPSC-CMs. As a safety concern, this important finding demonstrates that Neu5Gc can incorporate into hiPSC-CMs during the differentiation process from xenogeneic substances in the XC culture media.
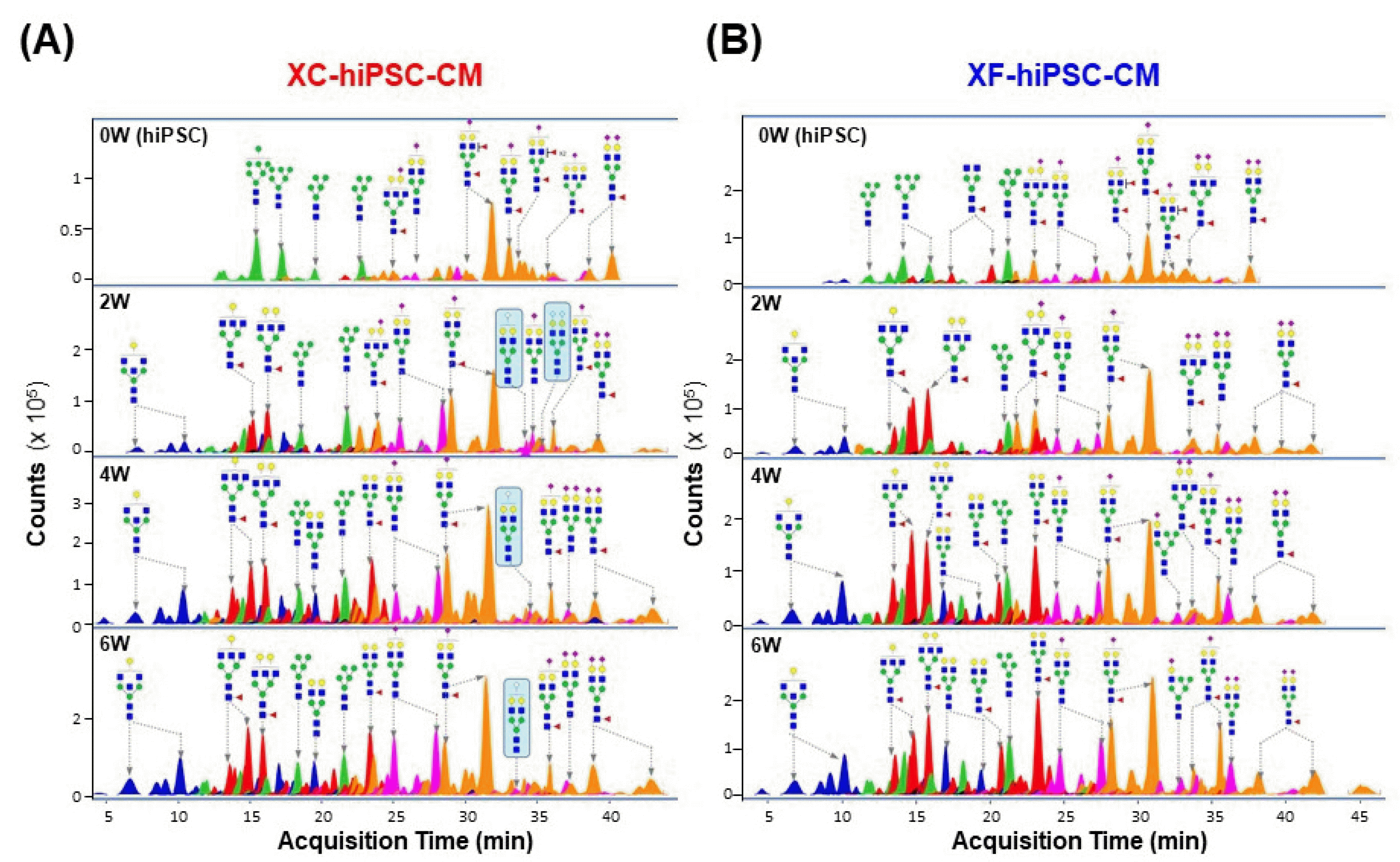
To confirm the increase in the Neu5Gc content in XC-hiPSC-CM during differentiation, we further quantified Neu5Gc and Neu5Ac in hiPSC-CM during the time course of differentiation (0, 2, 4, and 6 weeks) using liquid chromatography-multiple reaction monitoring-mass spectrometry (LC/MRM MS). In both XC- and XF-hiPSC-CM, the levels of Neu5Ac, which is abundant in both humans and other mammals, were significantly higher at 2 to 6 weeks of differentiation than at 0 weeks (hiPSC) (Fig. 5). The total sialic acid content was calculated as the sum of the Neu5Ac and Neu5Gc contents. Neu5Gc was detected after 2 weeks of differentiation (>0.6 ng/μg protein) and maintained the amount during the differentiation of cells into XC-hiPSC-CM by 4 weeks (Fig. 5A, upper right panel). In contrast, Neu5Gc was not detected in XF-hiPSC-CM at any time point (Fig. 5B, lower right panel). To confirm whether Neu5Gc present in the XC differentiation media was incorporated at the terminal position of N-linked glycans in hiPSC-CM, we further analyzed the N-linked glycan profiles of hiPSC-CM. Terminal Neu5Gc was detected on the glycan residues, which were separated from XC-hiPSC-CM during 2 to 6 weeks of differentiation. Neu5Gc was not detected in the glycan residues of XF-hiPSC-CM (Fig. 6). In this experiment using LC/MRM MS, the results of specific quantification of Neu5Ac/Neu5Gc confirmed the results of Neu5Gc detection by ELISA analysis. Moreover, from the results of glycan profiling analysis, the quantified amounts of Neu5Gc are derived from the XC-hiPSC-CM during the differentiation process, but not from the media itself.
This study is the first to demonstrate that the presence of xenogeneic substances in culture media can affect the structural maturation and functional properties of CM during the biomanufacturing of components used in cell-based therapies, especially in the differentiation of hiPSC-CM from hiPSC. We performed a comparative analysis of the characteristics of XF- and XC-hiPSC-CM, assessing gene and protein expression and electrophy-siological beating activity. Using a modified indirect ELISA, we also established a robust and reliable method for detecting Neu5Gc in XC-hiPSC-CM lysates without cross-reaction with Neu5Ac. The results of Neu5Gc detection using indirect ELISA were confirmed by the quantification of Neu5Gc in hiPSC-CM using LC/MRM MS. Based on our findings, we suggest the use of Neu5Gc, a xenogeneic sialic acid owing to a defect in the gene encoding the Neu5Gc biosynthetic enzyme CMAH in humans, as a potential surrogate marker for unexpected xenosialitis from cell-based biopharmaceuticals in patients.
Reportedly, biopharmaceuticals produced in the presence of animal-derived substances can induce xenogeneic immune responses (6). MatrigelTM, which is derived from mouse sarcoma cells, is frequently used as a material in scaffold matrices for the effective attachment and differentiation of CM (5, 27, 28). To avoid the potential risks of tumorigenicity associated with MatrigelTM, we successfully established a VTN-N-based XF differentiation method for hiPSC-CM using E8 media, which does not contain any animal-derived substances (29). The differentiation of hiPSC into hiPSC-CM in the VTN-N-based XF differentiation media induced rapid and synchronized beating activities in the CM, as compared to that achieved with MatrigelTM-based XC differentiation. It has been reported that stem cell expandability and its differentiation can be affected by components of culture media and scaffold, specifically in human stem cells (30, 31), and hiPSC-CM (32, 33). Our findings also suggest the importance of selecting appropriate scaffold matrix materials during differentiation.
To evaluate the maturation of XC- and XF-hiPSC-CM, we assessed the maturation parameters of CM, including the contractile cytoskeleton morphology, CM beating activity, electrophysiological action potential, and gene expression during calcium handling (3, 34). Even though mature CM that formed from XC-hiPSC-CM and XF-hiPSC-CM showed similar characteristics at 4 to 6 weeks of differentiation, XF-hiPSC-CM showed a higher expression of cardiac lineage-specific marker genes (MYH7, ACTC1, and TNNT2) and more rapid electrophysiological beating activity than XC-hiPSC-CM at 2 to 4 weeks of differentiation, which indicates the early onset of CM-oriented maturation in these cells. The maturation of human embryonic stem cell-derived CM is accompanied by changes in ion channel expression, with relevant electrophysio-logical consequences (35). RyR2, a large calcium ion channel, is frequently used as a marker for CM maturation since RyR2 accelerates membrane depolarization by releasing Ca+2 during cardiac muscle contraction (36, 37). In this study, RYR2 expression was relatively higher in XF-hiPSC-CM than in XC-hiPSC-CM at 2 weeks of differentiation, showing consistency with the contractile parameter (BPM and FPDcF) data obtained from the MEA assay. The BPM and FPDcF at 2 weeks were higher in XF-hiPSC-CM than in XC-hiPSC-CM, and morphological changes followed by CM beating activity also appeared more rapidly in XF-hiPSC-CM. Although the molecular mechanisms for the faster maturity of XF-hiPSC-CM than XC-hiPSC-CM remain unclear, we speculate that one of the potential mechanisms would be involved in the difference of glycosylation profiles during differentiation and maturation of hiPSC-CM. However, for clarification of the mechanism, further study would be needed.
A major concern associated with cell-based therapies is the unintended induction of immune responses. The use of common animal-derived substances, such as FBS, porcine trypsin, and bovine serum albumin, in cultures is associated with the potential risk of immune response induction in patients. In this study, we first demonstrated that Neu5Gc can be incorporated into cells from culture media during hiPSC-CM differentiation, as observed using an indirect ELISA and LC/MRM MS. A high Neu5Gc content was observed in the XC media and XC-hiPSC-CM but not in the XF media or XF-hiPSC-CM. Although it is challenging to determine the absolute Neu5Gc content in humans, the quantification of Neu5Gc in human serum using label-free ZIC-HILIC/MRM MS has been reported (21). Here, we successfully demonstrated the comprehensive quantification of Neu5Gc in hiPSC-CM during differentiation in the presence/absence of xenogeneic substances using LC/MRM MS. In a previous study, undiffe-rentiated stem cells were shown to contain glycans with high levels of mannose residues, whereas glycopeptides containing high levels of sialic acid were found in late-stage differentiated cells (19). To confirm that Neu5Gc quantified in hiPSC-CM was derived from the XC differentiation media, we further analyzed the glycan profiles of cell membrane glycoproteins in hiPSC-CM. Glycans with the Neu5Gc terminal sialic acid were detected in XC-hiPSC-CM lysates between 2 and 6 weeks of differentiation, but not in XC-hiPSC, demonstrating the incorporation of Neu5Gc from XC media into the glycans in the XC-hiPSC-CM membrane.
There are some limitations to this study. First, this study was unable to show the inflammatory gene or protein expressions in XC-hiPSC-CM although Neu5Gc is suggested as a potential surrogate marker for xenosialitis from cell-based biotherapeutics. Second, galactose-α-1,3-galactose (α-Gal) oligosaccharide is another xenogeneic substance in biologics that is present in all mammals except for humans and old-world nonhuman primates (38). α-Gal, the most abundant and hyperacute rejection-related xenogeneic antigen, can cause acute anaphylaxis in patients (39-41); thus, it should be evaluated in hiPSC-CM during the biomanufacturing process using proper detection and quantification methods in future studies.
In conclusion, our findings demonstrate that differentiation in the presence of xenogeneic substances can affect the functional maturation of hiPSC-CM and the potential occurrence of xenosialitis in patients. We also suggest the use of Neu5Gc as a surrogate marker for xenosialitis potentially induced in response to cell-based the-rapies. Taken together, the establishment of an optimized XF-biomanufacturing process with the verification of the xenogeneic immune response is essential for quality control and safety measures based on the regulatory aspects of cell-based biotherapeutics.
Notes
Author Contributions
Conceptualization: M Park, YG Kim, and JH Yun; Experiments and data analysis: YG Kim, JH Yun, JW Park, H-A Lee, and D Seong; Management support: S Lee, KD Park; Writing - original draft preparation: YG Kim, JH Yun, JW Park, and H-A Lee; Writing - review and editing: M Park.
Supplementary Materials
Supplementary data including one table, two figures, and one video can be found with this article online at https://doi.org/10.15283/ijsc22158.
References
1. Tsao CW, Aday AW, Almarzooq ZI, Alonso A, Beaton AZ, Bittencourt MS, Boehme AK, Buxton AE, Carson AP, Commodore-Mensah Y, Elkind MSV, Evenson KR, Eze-Nliam C, Ferguson JF, Generoso G, Ho JE, Kalani R, Khan SS, Kissela BM, Knutson KL, Levine DA, Lewis TT, Liu J, Loop MS, Ma J, Mussolino ME, Navaneethan SD, Perak AM, Poudel R, Rezk-Hanna M, Roth GA, Schroeder EB, Shah SH, Thacker EL, VanWagner LB, Virani SS, Voecks JH, Wang NY, Yaffe K, Martin SS. 2022; Heart Disease and Stroke Statistics-2022 update: a report from the American Heart Association. Circulation. 145:e153–e639. DOI: 10.1161/CIR.0000000000001052. PMID: 35078371.

2. Liu YW, Chen B, Yang X, Fugate JA, Kalucki FA, Futaku-chi-Tsuchida A, Couture L, Vogel KW, Astley CA, Balde-ssari A, Ogle J, Don CW, Steinberg ZL, Seslar SP, Tuck SA, Tsuchida H, Naumova AV, Dupras SK, Lyu MS, Lee J, Hailey DW, Reinecke H, Pabon L, Fryer BH, MacLellan WR, Thies RS, Murry CE. 2018; Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat Biotechnol. 36:597–605. DOI: 10.1038/nbt.4162. PMID: 29969440. PMCID: PMC6329375.

3. Karbassi E, Fenix A, Marchiano S, Muraoka N, Nakamura K, Yang X, Murry CE. 2020; Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine. Nat Rev Cardiol. 17:341–359. DOI: 10.1038/s41569-019-0331-x. PMID: 32015528. PMCID: PMC7239749.

4. Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, Gantz JA, Fugate JA, Muskheli V, Gough GM, Vogel KW, Astley CA, Hotchkiss CE, Baldessari A, Pabon L, Reinecke H, Gill EA, Nelson V, Kiem HP, Laflamme MA, Murry CE. 2014; Human embryonic-stem-cell-derived cardiomyocytes rege-nerate non-human primate hearts. Nature. 510:273–277. DOI: 10.1038/nature13233. PMID: 24776797. PMCID: PMC4154594.

5. Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. 2012; Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 109:E1848–E1857. DOI: 10.1073/pnas.1200250109. PMID: 22645348. PMCID: PMC3390875.

6. Martin MJ, Muotri A, Gage F, Varki A. 2005; Human embryonic stem cells express an immunogenic nonhuman sialic acid. Nat Med. 11:228–232. DOI: 10.1038/nm1181. PMID: 15685172.

7. Herberts CA, Kwa MS, Hermsen HP. 2011; Risk factors in the development of stem cell therapy. J Transl Med. 9:29. DOI: 10.1186/1479-5876-9-29. PMID: 21418664. PMCID: PMC3070641.

8. Lin Y, Zou J. 2020; Differentiation of cardiomyocytes from human pluripotent stem cells in fully chemically defined conditions. STAR Protoc. 1:100015. DOI: 10.1016/j.xpro.2020.100015. PMID: 32734277. PMCID: PMC7392178.

9. Ashok P, Parikh A, Du C, Tzanakakis ES. 2020; Xenogeneic-free system for biomanufacturing of cardiomyocyte progeny from human pluripotent stem cells. Front Bioeng Biotech-nol. 8:571425. DOI: 10.3389/fbioe.2020.571425. PMID: 33195131. PMCID: PMC7644809.

10. Chou HH, Takematsu H, Diaz S, Iber J, Nickerson E, Wright KL, Muchmore EA, Nelson DL, Warren ST, Varki A. 1998; A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci U S A. 95:11751–11756. DOI: 10.1073/pnas.95.20.11751. PMID: 9751737. PMCID: PMC21712.

11. Irie A, Koyama S, Kozutsumi Y, Kawasaki T, Suzuki A. 1998; The molecular basis for the absence of N-glycolylneuraminic acid in humans. J Biol Chem. 273:15866–15871. DOI: 10.1074/jbc.273.25.15866. PMID: 9624188.

12. Varki A, Schnaar RL, Schauer R. Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, Seeberger PH, editors. 2015. Sialic acids and other nonulosonic acids. Essentials of Glycobiology. 3rd ed. Cold Spring Harbor Laboratory Press;Cold Spring Harbor: p. 179–195.
13. Tangvoranuntakul P, Gagneux P, Diaz S, Bardor M, Varki N, Varki A, Muchmore E. 2003; Human uptake and incorpo-ration of an immunogenic nonhuman dietary sialic acid. Proc Natl Acad Sci U S A. 100:12045–12050. DOI: 10.1073/pnas.2131556100. PMID: 14523234. PMCID: PMC218710.

14. Hedlund M, Padler-Karavani V, Varki NM, Varki A. 2008; Evidence for a human-specific mechanism for diet and antibody-mediated inflammation in carcinoma progression. Proc Natl Acad Sci U S A. 105:18936–18941. DOI: 10.1073/pnas.0803943105. PMID: 19017806. PMCID: PMC2596253.

15. Bardor M, Nguyen DH, Diaz S, Varki A. 2005; Mechanism of uptake and incorporation of the non-human sialic acid N-glycolylneuraminic acid into human cells. J Biol Chem. 280:4228–4237. DOI: 10.1074/jbc.M412040200. PMID: 15557321.

16. Padler-Karavani V, Hurtado-Ziola N, Pu M, Yu H, Huang S, Muthana S, Chokhawala HA, Cao H, Secrest P, Friedmann-Morvinski D, Singer O, Ghaderi D, Verma IM, Liu YT, Messer K, Chen X, Varki A, Schwab R. 2011; Human xeno-autoantibodies against a non-human sialic acid serve as novel serum biomarkers and immunotherapeutics in cancer. Cancer Res. 71:3352–3363. DOI: 10.1158/0008-5472.CAN-10-4102. PMID: 21505105. PMCID: PMC3085609.

17. Samraj AN, Pearce OM, Läubli H, Crittenden AN, Bergfeld AK, Banda K, Gregg CJ, Bingman AE, Secrest P, Diaz SL, Varki NM, Varki A. 2015; A red meat-derived glycan promotes inflammation and cancer progression. Proc Natl Acad Sci U S A. 112:542–547. DOI: 10.1073/pnas.1417508112. PMID: 25548184. PMCID: PMC4299224.

18. Bashir S, Fezeu LK, Leviatan Ben-Arye S, Yehuda S, Reuven EM, Szabo de Edelenyi F, Fellah-Hebia I, Le Tourneau T, Imbert-Marcille BM, Drouet EB, Touvier M, Roussel JC, Yu H, Chen X, Hercberg S, Cozzi E, Soulillou JP, Galan P, Padler-Karavani V. 2020; Association between Neu5Gc carbohydrate and serum antibodies against it provides the molecular link to cancer: French NutriNet-Santé study. BMC Med. 18:262. DOI: 10.1186/s12916-020-01721-8. PMID: 32962714. PMCID: PMC7510162.

19. Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. 2013; Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 8:162–175. DOI: 10.1038/nprot.2012.150. PMID: 23257984. PMCID: PMC3612968.

20. Lee SJ, Kim HA, Kim SJ, Lee HA. 2021; Improving generation of cardiac organoids from human pluripotent stem cells using the aurora kinase inhibitor ZM447439. Biomedicines. 9:1952. DOI: 10.3390/biomedicines9121952. PMID: 34944767. PMCID: PMC8698385.
21. Seo N, Ko J, Lee D, Jeong H, Oh MJ, Kim U, Lee DH, Kim J, Choi YJ, An HJ. 2021; In-depth characterization of non-human sialic acid (Neu5Gc) in human serum using label-free ZIC-HILIC/MRM-MS. Anal Bioanal Chem. 413:5227–5237. DOI: 10.1007/s00216-021-03495-1. PMID: 34235565.

22. Hua S, Saunders M, Dimapasoc LM, Jeong SH, Kim BJ, Kim S, So M, Lee KS, Kim JH, Lam KS, Lebrilla CB, An HJ. 2014; Differentiation of cancer cell origin and molecular subtype by plasma membrane N-glycan profiling. J Proteome Res. 13:961–968. DOI: 10.1021/pr400987f. PMID: 24303873. PMCID: PMC3946297.

23. Oh MJ, Hua S, Kim BJ, Jeong HN, Jeong SH, Grimm R, Yoo JS, An HJ. 2013; Analytical platform for glycomic characterization of recombinant erythropoietin biotherapeutics and biosimilars by MS. Bioanalysis. 5:545–559. DOI: 10.4155/bio.12.327. PMID: 23425271.

24. Mummery CL, Zhang J, Ng ES, Elliott DA, Elefanty AG, Kamp TJ. 2012; Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ Res. 111:344–358. DOI: 10.1161/CIRCRESAHA.110.227512. PMID: 22821908. PMCID: PMC3578601.

25. Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. 2014; CellNet: network biology applied to stem cell engineering. Cell. 158:903–915. DOI: 10.1016/j.cell.2014.07.020. PMID: 25126793. PMCID: PMC4233680.

26. Jung G, Fajardo G, Ribeiro AJ, Kooiker KB, Coronado M, Zhao M, Hu DQ, Reddy S, Kodo K, Sriram K, Insel PA, Wu JC, Pruitt BL, Bernstein D. 2016; Time-dependent evolution of functional vs. remodeling signaling in induced pluripotent stem cell-derived cardiomyocytes and induced maturation with biomechanical stimulation. FASEB J. 30:1464–1479. DOI: 10.1096/fj.15-280982. PMID: 26675706. PMCID: PMC4799510.

27. Ojala M, Rajala K, Pekkanen-Mattila M, Miettinen M, Huhtala H, Aalto-Setälä K. 2012; Culture conditions affect cardiac differentiation potential of human pluripotent stem cells. PLoS One. 7:e48659. DOI: 10.1371/journal.pone.0048659. PMID: 23119085. PMCID: PMC3485380.

28. Feaster TK, Cadar AG, Wang L, Williams CH, Chun YW, Hempel JE, Bloodworth N, Merryman WD, Lim CC, Wu JC, Knollmann BC, Hong CC. 2015; Matrigel mattress: a method for the generation of single contracting human-induced pluripotent stem cell-derived cardiomyocytes. Circ Res. 117:995–1000. DOI: 10.1161/CIRCRESAHA.115.307580. PMID: 26429802. PMCID: PMC4670592.
29. Lim JJ, Kim HJ, Rhie BH, Lee MR, Choi MJ, Hong SH, Kim KS. 2019; Maintenance of hPSCs under Xeno-free and che-mically defined culture conditions. Int J Stem Cells. 12:484–496. DOI: 10.15283/ijsc19090. PMID: 31658510. PMCID: PMC6881038.
30. Hua Y, Yoshimochi K, Li J, Takekita K, Shimotsuma M, Li L, Qu X, Zhang J, Sawa Y, Liu L, Miyagawa S. 2022; Development and evaluation of a novel xeno-free culture medium for human-induced pluripotent stem cells. Stem Cell Res Ther. 13:223. DOI: 10.1186/s13287-022-02879-z. PMID: 35658933. PMCID: PMC9166585.

31. Tateishi K, Ando W, Higuchi C, Hart DA, Hashimoto J, Nakata K, Yoshikawa H, Nakamura N. 2008; Comparison of human serum with fetal bovine serum for expansion and differentiation of human synovial MSC: potential feasibility for clinical applications. Cell Transplant. 17:549–557. DOI: 10.3727/096368908785096024. PMID: 18714674.
32. Sung TC, Liu CH, Huang WL, Lee YC, Kumar SS, Chang Y, Ling QD, Hsu ST, Higuchi A. 2019; Efficient differentiation of human ES and iPS cells into cardiomyocytes on biomaterials under xeno-free conditions. Biomater Sci. 7:5467–5481. DOI: 10.1039/C9BM00817A. PMID: 31656967.
33. Villa-Diaz LG, Ross AM, Lahann J, Krebsbach PH. 2013; Concise review: the evolution of human pluripotent stem cell culture: from feeder cells to synthetic coatings. Stem Cells. 31:1–7. DOI: 10.1002/stem.1260. PMID: 23081828. PMCID: PMC3537180.

34. Henderson CA, Gomez CG, Novak SM, Mi-Mi L, Gregorio CC. 2017; Overview of the muscle cytoskeleton. Compr Physiol. 7:891–944. DOI: 10.1002/cphy.c160033. PMID: 28640448. PMCID: PMC5890934.

35. Bosman A, Sartiani L, Spinelli V, Del Lungo M, Stillitano F, Nosi D, Mugelli A, Cerbai E, Jaconi M. 2013; Molecular and functional evidence of HCN4 and caveolin-3 interaction during cardiomyocyte differentiation from human embryonic stem cells. Stem Cells Dev. 22:1717–1727. DOI: 10.1089/scd.2012.0247. PMID: 23311301. PMCID: PMC3657289.

36. Medeiros-Domingo A, Bhuiyan ZA, Tester DJ, Hofman N, Bikker H, van Tintelen JP, Mannens MM, Wilde AA, Ackerman MJ. 2009; The RYR2-encoded ryanodine receptor/calcium release channel in patients diagnosed previously with either catecholaminergic polymorphic ventricular tachycardia or genotype negative, exercise-induced long QT syndrome: a comprehensive open reading frame mutational analysis. J Am Coll Cardiol. 54:2065–2074. DOI: 10.1016/j.jacc.2009.08.022. PMID: 19926015. PMCID: PMC2880864.

37. Yang HT, Tweedie D, Wang S, Guia A, Vinogradova T, Bogdanov K, Allen PD, Stern MD, Lakatta EG, Boheler KR. 2002; The ryanodine receptor modulates the spontaneous beating rate of cardiomyocytes during development. Proc Natl Acad Sci U S A. 99:9225–9230. DOI: 10.1073/pnas.142651999. PMID: 12089338. PMCID: PMC123122.

38. Galili U. 1999; Evolution of alpha 1,3galactosyltransferase and of the alpha-Gal epitope. Subcell Biochem. 32:1–23. DOI: 10.1007/978-1-4615-4771-6_1. PMID: 10391989.
39. Chung CH, Mirakhur B, Chan E, Le QT, Berlin J, Morse M, Murphy BA, Satinover SM, Hosen J, Mauro D, Slebos RJ, Zhou Q, Gold D, Hatley T, Hicklin DJ, Platts-Mills TA. 2008; Cetuximab-induced anaphylaxis and IgE specific for galactose-alpha-1,3-galactose. N Engl J Med. 358:1109–1117. DOI: 10.1056/NEJMoa074943. PMID: 18337601. PMCID: PMC2361129.

40. Swiontek K, Morisset M, Codreanu-Morel F, Fischer J, Mehlich J, Darsow U, Petitpain N, Biedermann T, Ollert M, Eberlein B, Hilger C. 2019; Drugs of porcine origin- a risk for patients with α-gal syndrome? J Allergy Clin Immunol Pract. 7:1687–1690.e3. DOI: 10.1016/j.jaip.2018.12.005. PMID: 30557715.
41. Chinuki Y, Morita E. 2019; Alpha-Gal-containing biologics and anaphylaxis. Allergol Int. 68:296–300. DOI: 10.1016/j.alit.2019.04.001. PMID: 31053502.

Fig. 1
Schematic representation of the protocol for the differentiation of hiPSC into CM. (A) In the XC culture condition, hiPSC-CM were generated on MatrigelTM-coated plates according to the modified GiAB (MM.1) protocol. Up to 5 days after the differentiation into CM commenced, 1~1.5 vol% MatrigelTM was added to the medium. (B) In the XF culture condition, hiPSC were differentiated into cardiac-lineage cells on VTN-N-coated 12-well culture plates using the PSC Cardiomyocyte Differentiation Kit.
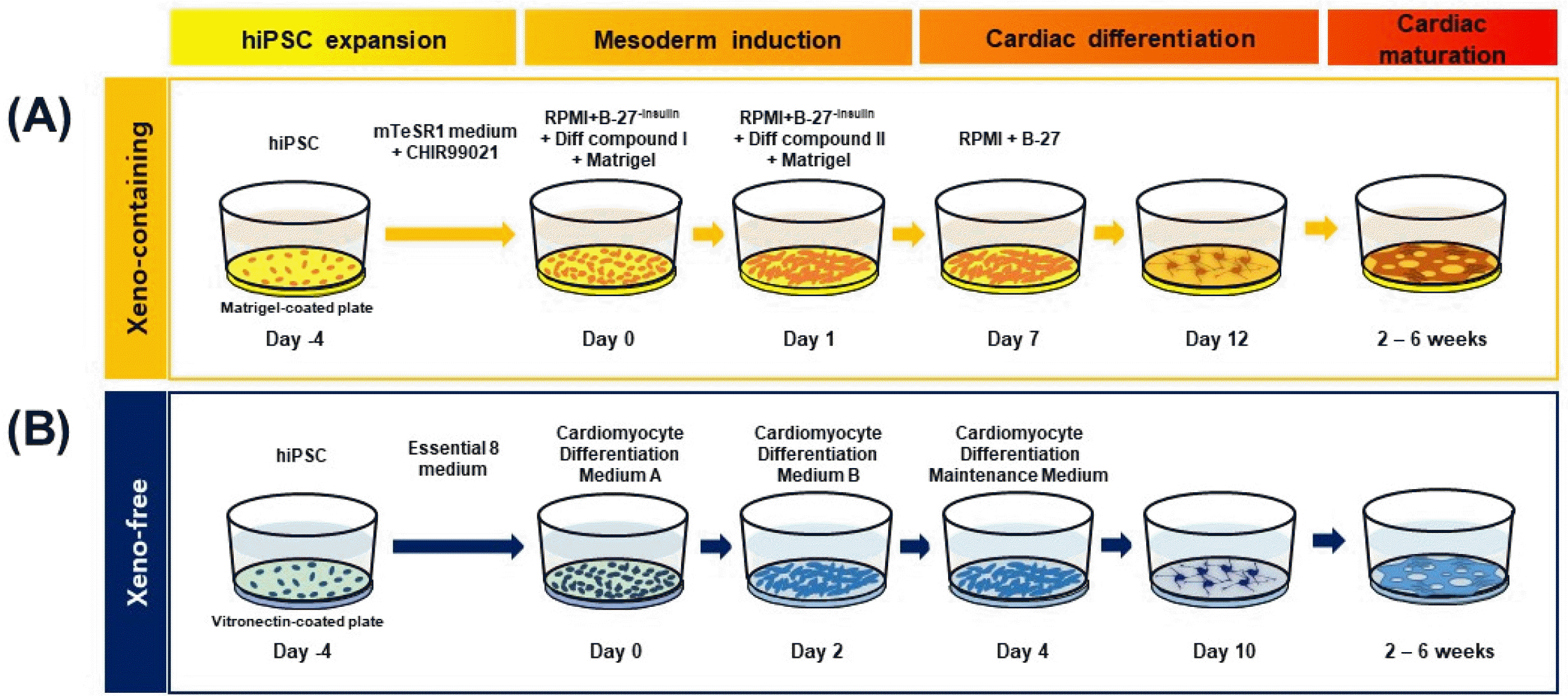
Fig. 2
Analysis of the characteristics of hiPSC-CM. (A) Microscopic image of cells of cardiac lineage at 1, 2, and 4 weeks after the differentiation of hiPSC into CM under XC and XF conditions. (B) mRNA expressions of cardiac lineage-specific marker genes in XC- and XF-hiPSC-CM were compared at 0, 2, and 4 weeks post-differentiation using reverse transcription-quantitative PCR. The expression values were normalized against that of 18S rRNA as a housekeeping gene. n=3 per group, *p<0.05, XC-hiPSC-CM vs. XF-hiPSC-CM using unpaired Student’s t-test. (C) Protein expression of cardiac lineage-specific markers was determined by immunostaining at 4 weeks after differentiation under the XF condition. MLC-2v, myosin light chain-2v (ventricular/cardiac muscle isoform) (red); cTnT, cardiac troponin T (green); α-SMA, α-smooth muscle actin (red); α-actinin, a cytoskeletal actin-binding protein (green). Cell nuclei were counterstained using DAPI (blue).
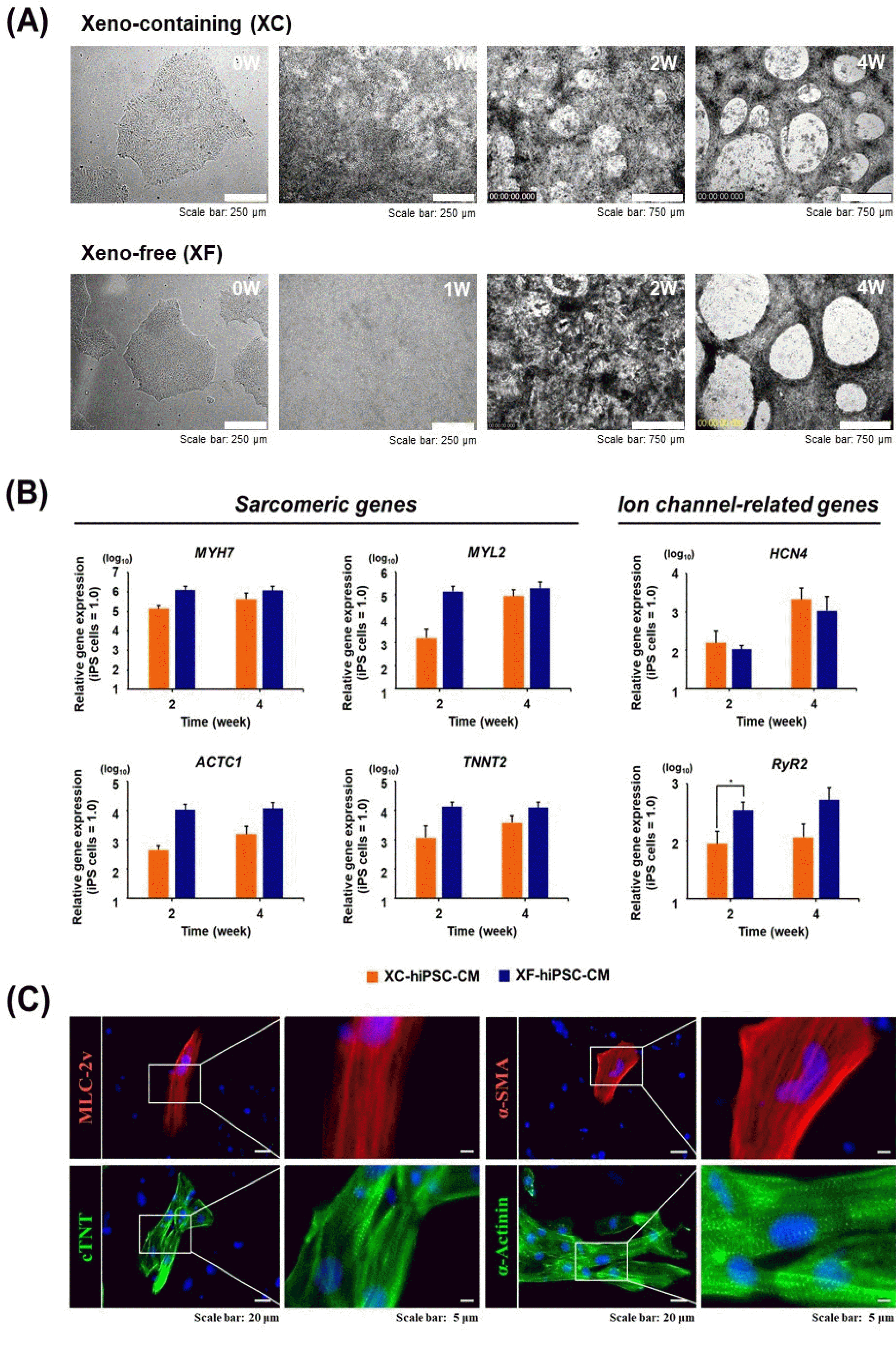
Fig. 3
Contractile properties of CM that differentiated from hiPSC. The electrophysiological properties of hiPSC-CM were assessed using a multi-electrode array (MEA) assay. Differences in the field potential parameters of (A) XC-hiPSC-CM and (B) XF-hiPSC-CM at 2 and 4 weeks after differentiation. *p<0.05, **p<0.01, ***p<0.001. (C) Comparison of the field potential parameters of XC-hiPSC-CM and XF-hiPSC-CM at 4 weeks. The commercially available CM NexelTM Cardiosight®-S was used as a positive control. n=6 per group of XC/XF-CM, n=4 for NexelTM Cardiosight®-S. ***p<0.001 by one-way analysis of variance with Tukey’s multiple comparisons test. Val-ues are presented in terms of means± SEM. BPM, beats per minute; FPA, field potential amplitude; FPDcF, field potential duration corrected with Fride-ricia's formula.
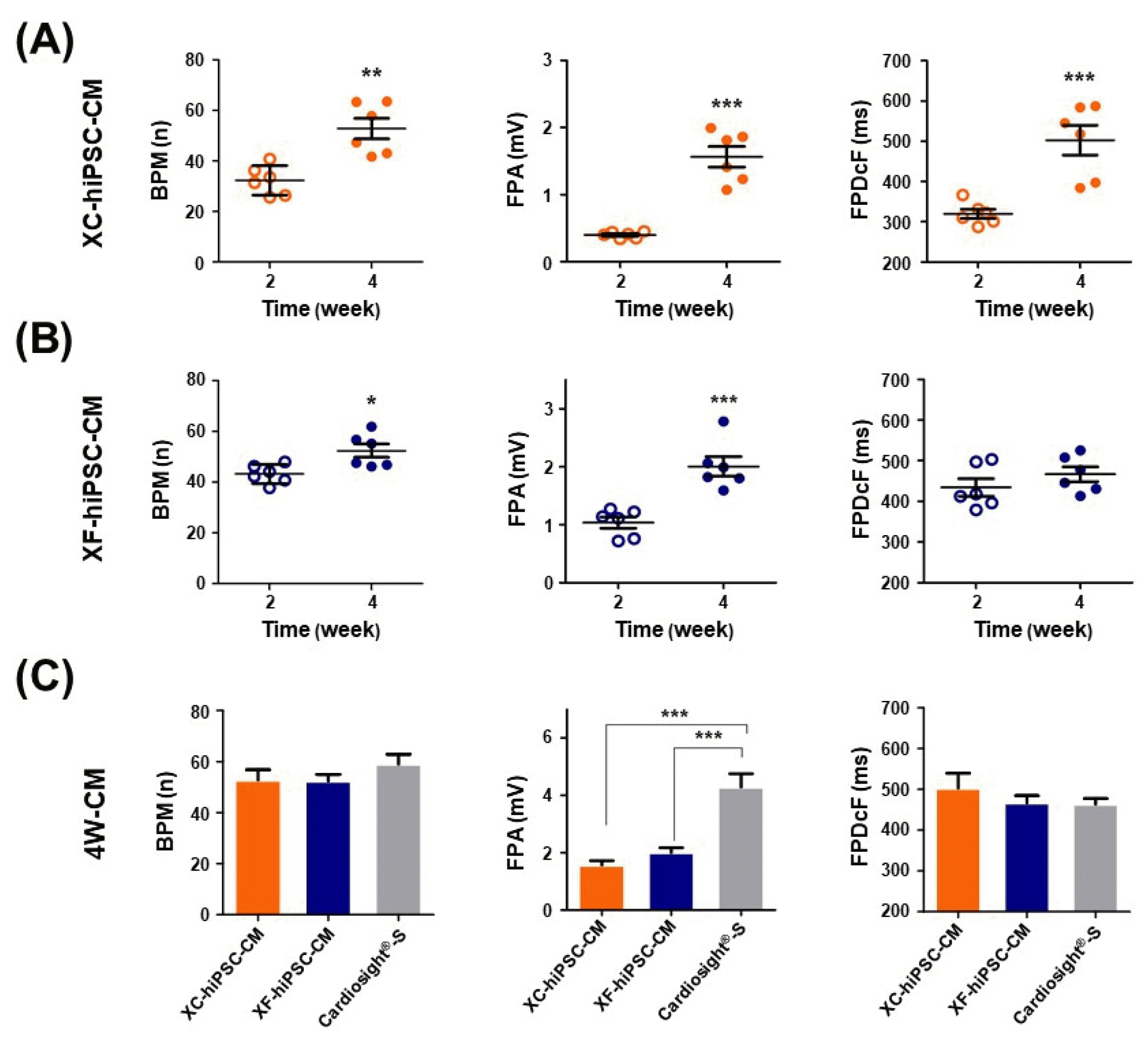
Fig. 4
Evaluation of immunogenic Neu5Gc in hiPSC-CM using an indirect ELISA. (A) Neu5Gc was only detected in XC complete media. (B) Neu5Gc expression in the XC-conditioned media and XC-hiPSC-CM lysates was significantly higher at 2 to 4 weeks of differentiation than at 0 weeks (hiPSC). Neu5Gc was not detected in XF media and XF-hiPSC-CM lysates. Fetal bovine serum was used as a positive control and experimental internal control. Statistical analysis was performed using analysis of variance, followed by Tukey’s multiple comparisons test. Values are presented in terms of means±SEM. **p<0.01, ***p<0.001. n=3 per group.
Fig. 5
Quantification of Neu5Gc in the hiPSC-CM lysate using LC/MRM MS. The total sialic acid content (Ne-u5Ac and Neu5Gc) was significantly higher at 2 to 6 weeks (W) after differentiation into hiPSC-CM than at 0 weeks (hiPSCs) under both (A) XC condition and (B) XF conditions. Neu5Gc was only detected in XC-hiPSC-CM, even though the most prominent increase in the level of sialic acid was attributed to Neu5Ac. Statistical analysis was performed using the Student’s t-test. *p<0.05, ***p<0.001. Values are presented in terms of means±SEM. n=3 per group. N.D., not detected.

Fig. 6
Sialic acid residues on the N-linked glycans of hiPSC-CM. The N-linked glycan structures are indicated by each major peak with the acquisition time. (A) Terminal Neu5Gc was detected on the glycans that were separated from XC-hiPSC-CM during 2 to 6 weeks of differentiation (indicated in the blue box). (B) Neu5Gc was not detected in the glycan residues of XF-hiPSC-CM. 0W, hiPSC; 2-6W, hiPSC-CM.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download