INTRODUCTION
Ralstonia spp. is a Gram-negative, non-fermentative, aerobic rod-shaped genus that includes
R. pickettii, R. insidiosa, R. solanacearum, and
R. mannitolilytica, formerly known as
Burkholderia pickettii, B. solanacearum,
Pseudomonas solanacearum, and
P. thomasii.
1 This genus is commonly found in water supplies, soil, and plants, and has the ability to survive in low-nutrient conditions and even in the presence of disinfectants.
2
Ralstonia spp. can produce biofilm, allowing it to persist in the environment, evade the host’s immune responses, and cause invasive diseases, particularly in immunocompromised patients, including those with hematologic malignancies, solid organ or hematopoietic stem cell transplants,
34 patients in intensive care or with central venous catheters (CVCs),
2356 patients with cystic fibrosis, and premature infants.
35789
R. mannitolilytica has been implicated in healthcare-associated infections such as central line-associated infection, pneumonia, and meningitis
3461011 and is primarily derived from water; however, outbreaks have also been linked to contaminated oxygen delivery devices or multi-dose bottles of saline.
37 In addition to contaminated solutions, contaminated disinfectants have also been considered as sources of
R. mannitolilytica infection.
2
Ralstonia spp. is resistant to various antibiotics, including extended generation cephalosporins, most aminoglycosides, fluoroquinolones, and carbapenems.
2591213 Although there are limited studies on the antibiotic resistance mechanisms of
R. mannitolilytica, it has been reported that this species contains class D beta-lactamases such as OXA-22 and OXA-60 in the chromosome, which are linked to intrinsic resistance.
131415 Whole-genome sequence analysis has revealed that one strain of
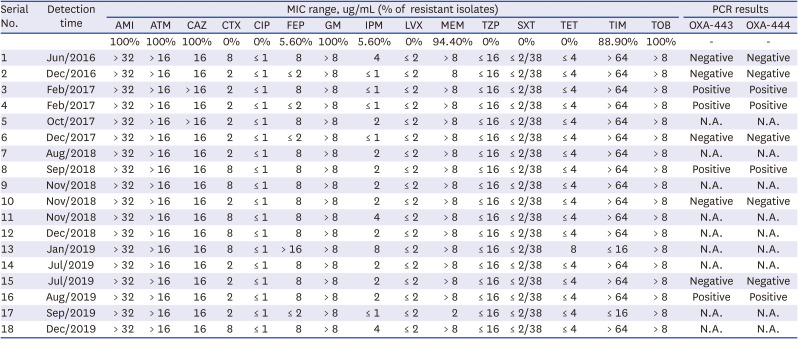
R. mannitolilytica carries OXA-443/OXA-22-like and OXA-444/OXA-60-like β-lactamase.
14 OXA-444/OXA-60-like β-lactamase is an extended-spectrum oxacillinase with carbapenem-hydrolyzing properties.
15
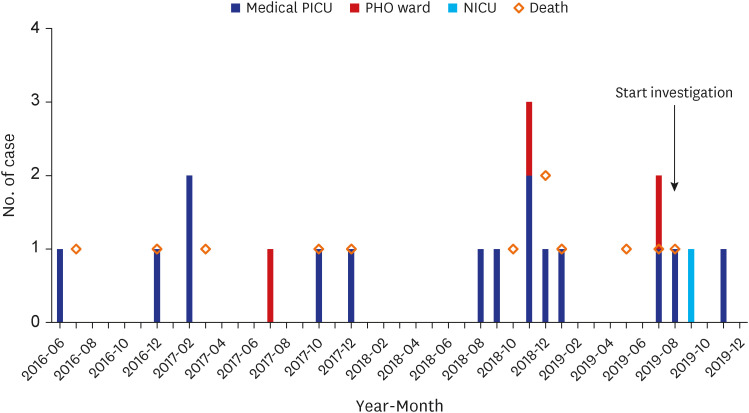
From August 2018 to November 2019, we detected several small clusters of mainly meropenem-resistant R. mannitolilytica infections, some of which resulted in fatalities. The objective of this study is to analyze the clinical characteristics of R. mannitolilytica infections in pediatric patients admitted to Asan Medical Center Children’s Hospital and to present an epidemiological investigation of the outbreak.
METHODS
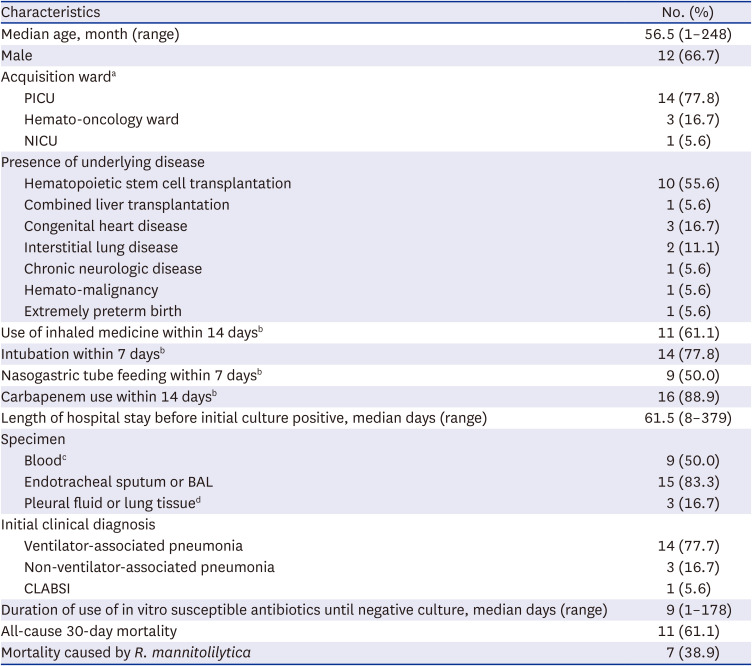
Patients with culture-confirmed
R. mannitolilytica infection and consistent signs of infection who were admitted to Asan Medical Center Children’s Hospital, in Seoul, South Korea from March 2009 to March 2023 were included in this study. Our center is a 270-bed tertiary referral center consisting of three general pediatric wards, one pediatric hemato-oncology (PHO) ward with 41 beds, three neonatal intensive care units (NICUs) with 58 beds, and two pediatric intensive care units (PICUs) with a total of 25 beds (including medical and surgical PICUs). Patients admitted to the hospital included those with hematopoietric stem cell transplants, solid organ transplants, as well as surgical conditions such as cardiac, neurosurgical, and orthopedic conditions. Clinical and demographic data were abstracted from electronic medical records, including age, sex, clinical diagnosis, underlying medical conditions,
R. mannitolilytica acquisition ward, duration of hospitalization before the initial positive culture, use of antibiotics, need for mechanical ventilation, use of inhaled medicine, and all-cause 30-day mortality. Clinical diagnoses such as ventilator-associated pneumonia (VAP), non-ventilator-associated pneumonia, and central line-associated bloodstream infections (CLABSIs) were categorized according to CDC definitions.
1617 Strains were considered to have been acquired at ward A if patients were infected within 48 hours of transferring from ward A to ward B.
Species identification and antibiotics susceptibility
Species identification and antimicrobial susceptibilitiy tests were performed using the MicroScan WalkAway 96 plus system and Neg Combo Panel Type 72 (Beckman Coulter, Brea, CA, USA).
The antibiotic susceptibility tests for
Ralstonia spp. included amikacin, aztreonam, ceftazidime, cefotaxime, ciprofloxacin, cefepime, gentamicin, imipenem, levofloxacin, meropenem, piperacillin/tazobactam, trimethoprim/sulfamethoxazole, tetracycline, ticarcillin/clavulanate, and tobramycin. The results were interpreted using Clinical and Laboratory Standards Institute (CLSI) criteria for
Pseudomonas aeruginosa ATCC
® 27853, as there are no CLSI susceptibility breakpoints available for
Ralstonia spp.18
Polymerase chain reaction (PCR) for OXA-443/OXA-444 genes
R. mannitolilytica isolates stored at −70°C were thawed and subcultured for PCR to detect the OXA-443 and OXA-444 genes. The following primers were used: OXA-443 Forward 5′-ATGACGAAACTCCGCCA-3′/OXA-443, Reverse 5′-AGGTGGGCTCGATCTTG-3′, and OXA-444 Forward 5′-ATGTTCTCTCGTTGGTC-3′/ OXA-444 Reverse 5′-TGCGGGTCGGACGGAGA-3′. A 20 µL reaction mixture comprising distilled water 12.2 µL, 10× PCR buffer 2 µL (containing Mg 2+), primer set 0.5 µL each, 2.5 mM dNTP mixture 1.6 µL, 5U/µL Taq DNA polymerase 0.2 µL, and 3 µL of DNA extract were used for bacterial PCR amplifications. After placing reaction tubes within the GeneAmp PCR System 9700 (Applied Biosystems, Foster, CA, USA), amplification began with an initial denaturation at 95°C for 5 minutes, 35 cycles of 95°C for 1 minute, 55°C for 30 seconds, and 72°C for 1 minute, followed by a final extension at 72°C for 5 minutes. Distilled water was used as a negative control.
Epidemiological investigation and environmental sampling
Between August and September 2019, an epidemiological investigation was conducted to determine the source of R. mannitolilytica infections in the PICU and to prevent further outbreaks, following the second outbreak that occurred in July 2019. Environmental sampling was performed in the PICU using sterile cotton swabs (DAE-HAN cotton tip, South Korea) to collect samples from ventilator circuits, bed railings, monitor surfaces, basins, and water purifiers. The cotton swabs were soaked in distilled water and rubbed across the environment, and the collected samples were inoculated into meropenem disc-inoculated selective tryptic soy broth medium. After cultivation, the medium was inoculated into McConkey agar once it became cloudy to begin subculture. After the bacteria were isolated, matrix-assisted laser desoroption/ionization time-of-flight (MALDI-TOF) was performed to identify them.
Ethics statement
The present study protocol was reviewed and approved by the Institutional Review Board of Asan Medical Center (approval No. 2020-1142). A waiver of prior consent was granted, given the retrospective nature of the study.
DISCUSSION
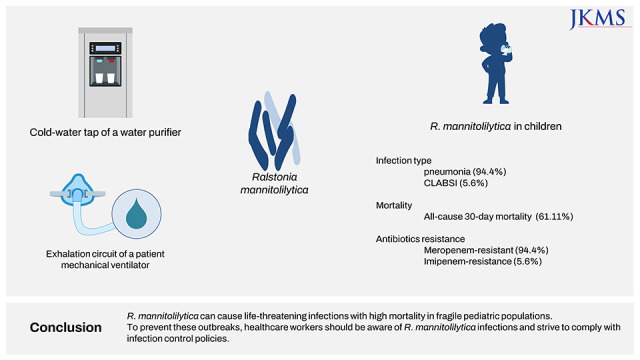
Since June 2016, when the first case of R. mannitolilytica infection was detected at our hospital, we have recorded a total of 18 pediatric cases with R. mannitolilytica infections. The 30-day all-cause mortality rate was as high as 61%, most of the cases were resistant to meropenem. Most patients were diagnosed with pneumonia, occurring as outbreaks in a medical PICU. Epidemiological investigations suggested that water purifiers might be the possible source of the outbreak.
Ralstonia spp. has emerged as an opportunistic pathogen in nosocomial settings.
123457911192021 While
Ralstonia spp. can cause invasive disease, most reports have shown a good prognosis and low mortality. For instance, during an outbreak in an oncology day ward, 22 patients with CVC developed bacteremia, which was suspected to be caused by flushing the CVCs with contaminated saline; however, symptoms improved in call cases after the removal of their CVCs.
3 Another report from a hemodialysis unit at a tertiary hospital recorded 16 cases of bacteremia outbreaks, with a high probability of dialysis water being the source. The mortality rate was 6%, and all cases were treated with antibiotics, including carbapenems, while dialysis catheters were changed except for one patient, who had their dialysis catheter locked with imipenem.
21 However, patients with cystic fibrosis may become chronically infected with this species, associated with severe respiratory infections and a mortality of up to 86%.
192223
Although there are few reports of
R. mannitolitytica infection in pediatric patients, an outbreak of
R. mannitolilytica bacteremia has been reported in neonates admitted to the NICU; however, the primary source of the outbreak was unknown.
9 Outbreaks associated with using a contaminated oxygen delivery device in pediatric patients have also been reported, leading to a national outbreak in the United States in 2005 and an outbreak in Israel in 2011.
720 Patients presented with respiratory infection and bacteremia, and all but one recovered. The one patient who did not recover was considered to have died of complicating underlying lung disease and not of
Ralstonia infection.
In our study, all cases of R. mannitolilytica infection occurred in pediatric patients with underlying serious medical conditions such as congenital heart disease, interstitial lung disease, chronic neurologic disease, hematopoietic stem cell transplantation recipients, and extremely preterm infants. In addition, 88.2% of cases occurred while staying in intensive care units. Our report suggests that pediatric patients with significant comorbidities who are infected with R. mannitolilytica infections may result in high mortality despite being treated with antibiotics that are susceptible in vitro. It is not clear why R. mannitolilytica infections have occurred only in pediatric patients and not in adult wards. However, considering that the first case of R. mannitolilytica infection in our hospital occurred in a child receiving long-term treatment in a PICU or PHO ward, it is believed that the strain responsible is not one that spreads widely. Nevertheless, continuous infection management and monitoring is crucial in preventing the spread of infections caused by this strain.
R. mannitolilytica has been found to modulate biofilm in plastic water pipes, and contaminated water supplies such as respiratory gas humidification devices and multi-dose saline bottles could be sources of
R. mannitolilytica infection.
3712
R. mannitolilytica could not be easily eradicated using disinfection protocols due to its ability to form biofilm structures that provide increased protection against external stress, such as disinfectants, and the ability to slow bacterial growth within the biofilm.
24
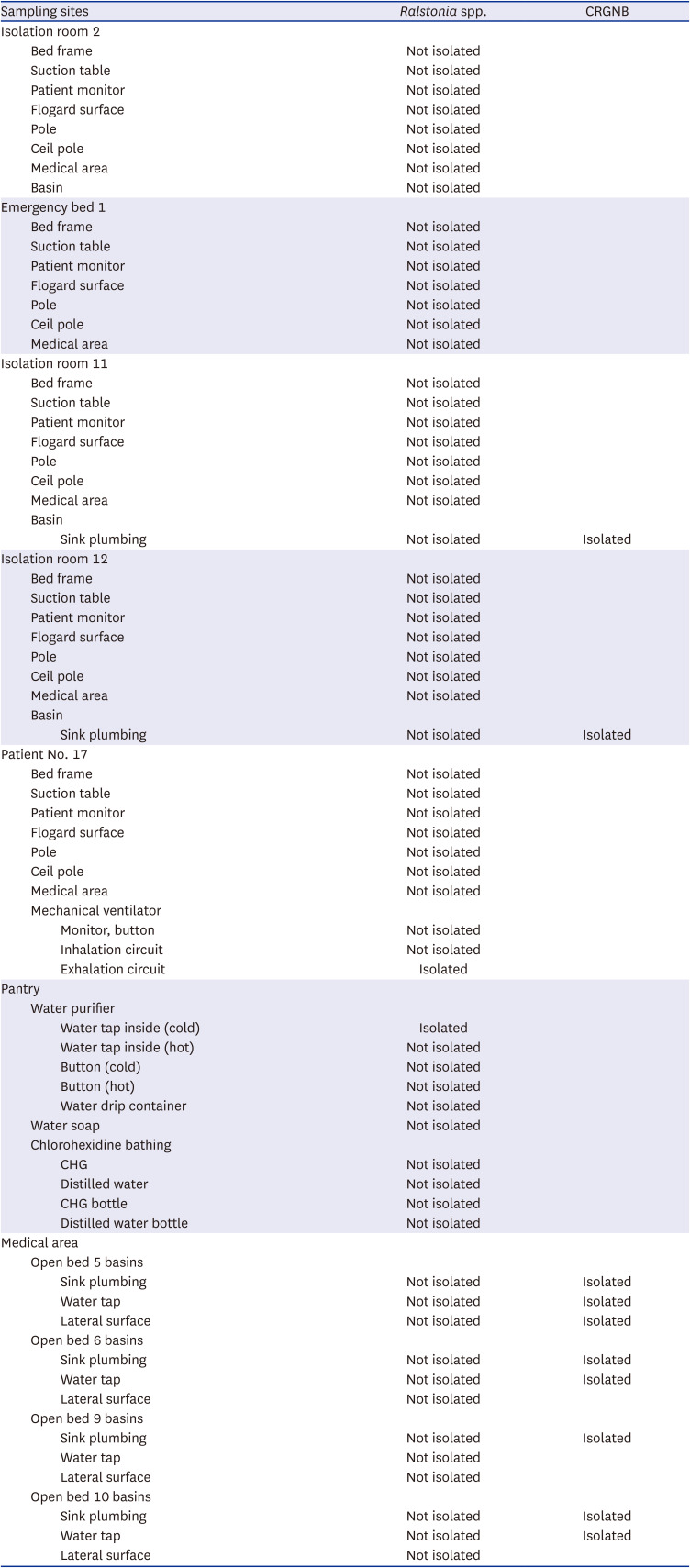
In this study, although environmental investigations were not conducted in locations other than the PICU, R. mannitolilytica was isolated from a water purifier in the pantry that was frequently used for drinking water for patients in the PICU. In addition, R. mannitolilytica was detected in the exhalation circuit of the ventilator but not in the inhalation circuit for one patient in the PICU, which suggest that it would be crucial to meticulously manage the exhalation circuit in patients receiving mechanical ventilation to minimize the potential spread of this pathogen.
Since 2018, our institute has utilized only single-dose bottles for inhaled medication, and there was no evidence indicating that the
R. mannitolilytica outbreak in 2019 was due to contaminated water used for inhaled medication or a contaminated circuit in the ventilator. A matched case-control study revealed that recovery of
R. mannitolilytica was not significantly associated with exposure to mechanical ventilation, nasogastric tube feeding, or inhaled medications. However,
R. mannitolilytica was isolated from the surface swabs of humidifier.
7 The exact source of the
R. mannitolilytica outbreak in 2019 could not be determined since pulsed-field gel electrophoresis or multilocus sequence typing was not performed on the strains isolated from the clinical specimens and environments, and the antibiotic susceptibility of
R. mannitolilytica isolates from environmental sampling, identified by MALDI-TOF, was not evaluated. Nevertheless, following a comprehensive approach that involved switching from a water purifier to bottled water, improving compliance with the VAP bundle, and emphasizing the importance of environmental cleanliness and hand hygiene, there were no further cases of
R. mannitolilytica infection reported until March 2023.
Although there is some variation in antibiotic resistance patterns among
R. mannitolilytica isolates in other studies, the species is known to be resistant to many different types of antibiotics; most isolates were resistant to gentamycin, tobramycin, colistin, aztreonam, meropenem or imipenem.
2345913
R. mannitolilytica isolates exhibited high MICs against meropenem compared with imipenem.
613 In this study, the resistance rate to meropenem and MIC against meropenem was also higher than imipenem. These findings may be partially explained by higher meropenem exposure, as overall meropenem usage accounted for a large pergentage of 96.5% of carbapenem use in our institute,
25 and most patients had a recent history of meropenem exposure in our study.
Although data on the mechanism of β-lactam, including carbapenem resistance in
Ralstonia spp. is limited, the presence of OXA β-lactamase is suspected to be the primary mechanism. Two chromosomally encoded class D β-lactamases, OXA-443 and OXA-444 (similar to OXA-22 and OXA-60, respectively, with 86.0% and 90.3% amino acid identities), have been reported as the primary mechanism for
R. mannitolilytica resistance.
314 OXA-22 is a narrow-spectrum oxacillinase, not an inducible carbapenemase, whereas OXA-60 is an inducible carbapenemase expressed only after
β-lactam induction (imipenem as a β-lactam inducer) and has imipenem-hydrolyzing properties.
15 The resistance genes extracted from the whole genome sequencing data showed that
R. mannitolilytica carried the resident narrow-spectrum oxacillinase
blaOXA-22 gene and intrinsically species-specific class D oxacillinase
blaOXA-443 and
blaOXA-444 genes, as well as the serin-hydrolase class C family β-lactamase.
5 However,
R. mannitolilytica did not carry carbapenemase genes (
blaKPC,
blaNDM,
blaOXA-48, and
blaVIM), according to RT-PCR (GeneXpert, Cepheid, USA).
5 The presence of OXA-444 and the serin-hydrolase class C β-lactamase, in association with different mechanisms such as porin deficiency or over-expression of efflux pumps, may confer resistance to multiple classes of antibiotics of
R. mannitolilytica.
3 In this study, all 18
R. mannitolilytica isolates were susceptible to cefotaxime and piperacillin/tazobactam, suggesting that they did not carry class C β-lactamase. Although only a small portion of isolates kept at −70°C and successfully subcultured could be used for the PCR of OXA-443 and OXA-444, there was no difference in antibiotic resistance patterns regardless of the presence of
OXA-443 and
OXA-444 genes. Further investigation focused on class D oxacillinase outside of
OXA-443 and
OXA-444 is required to elucidate the resistance mechanisms. The antimicrobial treatment of
R. mannitolilytica infection is challenging because
Ralstonia species are resistant to various antimicrobial agents, including carbapenems. Considering the antibiotic susceptibility of
R. mannitolilytica, initial empirical antibiotics should include piperacillin/tazobactam, quinolone, or trimethoprim/sulbactam, according to the clinical situation.
Our study has several limitations. Firstly, we could not accurately determine the souce of the R. mannitolilytica outbreaks due to the time lag to start environmental investigation from the first case. In addition, we did not evaluate molecular analysis including PFGE and antibiotic susceptibility in isolates obtained from environmental sampling, making it impossible to determine their consistency with patient isolates. Secondly, we only performed PCR on successfully sub-cultured strains and did not analyze genes other than OXA-443 and OXA-444 present in R. mannitolilytica. As a next study, whole genome analysis could thoroughly evaluate the genetic background of antibioitcs resistance of R. mannitolilytica. However, in this study, we reported clinical characteristics of the largest outbreak of R. mannitolilytica infections in Korean children and identified problems with the water supply as a potential soure. This finding highlights the significance of our study.
We reported an outbreak of R. mannitolilytica infection primarily presenting as pneumonia in children with underlying comorbidities. R. mannitolilytica can cause life-threatening infections with high mortality in vulnerable pediatric populations. To prevent these outbreaks, it is necessary to raise awareness about the significance of R. mannitolilytica infection and to comply with infection control policies, such as environmental cleaning and hand hygiene.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download