Abstract
Acute megakaryoblastic leukemia (AMKL) is a rare type of acute myeloid leukemia that evolved from primitive megakaryoblasts. We report a case of non-Down syndrome AMKL with CBFA2T3::GLIS2 fusion that has not been described before in Korea. A 20-month-old girl presented with a fever and was diagnosed with AMKL after a bone marrow study and flow cytometry analysis. An RNA fusion test observed a fusion transcript between exons 9 and 2 of the CBFA2T3 and GLIS2 genes, respectively. The patient achieved complete morphological remission after induction chemotherapy. However, her prognosis was poor as measurable residual disease remained, detected using reverse transcription PCR. RNA fusion analysis could be a helpful tool to identify clinically actionable genomic markers for patient risk stratification and measurable residual disease monitoring.
초록
급성거핵모구백혈병(AMKL)은 원시 거핵모구에서 유래한 드문 유형의 급성골수백혈병이다. 연구자들은 한국에서 한번도 보고되지 않은 CBFA2T3::GLIS2 유전자 융합을 가진 다운증후군이 아닌 AMKL의 사례를 보고하고자 한다. 발열을 주소로 내원한 20개월 여아가 골수 검사와 유세포분석을 통해서 AMKL로 진단되었다. RNA fusion panel 검사를 시행하여 CBFA2T3의 9번 엑손과 GLIS2의 2번 엑손 사이의 융합 전사체를 발견했다. 환자는 유도 화학요법을 투여 받은 후 형태학적 완전 관해를 보였지만, 역전사 중합효소연쇄반응으로 측정한 미세잔존질환은 양성을 보였고 나쁜 예후를 보였다. RNA fusion 검사는 환자의 위험도를 계층화하고 미세잔존질환을 추적관찰하는 등 임상적인 의미를 갖는 유전적 표지자를 검출하기 위한 유용한 도구로 사용될 수 있을 것으로 보인다.
Acute megakaryoblastic leukemia (AMKL) is a rare type of acute myeloid leukemia (AML) that evolved from primitive megakaryoblasts and is classified in the French-American-British Classification of AML as M7 [1]. AMKL is classified mainly as pediatric and adult cases, and pediatric cases are classified as Down syndrome AMKL (DS-AMKL) and non-Down syndrome AMKL (non-DS-AMKL).
The CBFA2T3::GLIS2 fusion, resulting from a cryptic inversion of chromosome 16, is identified in up to 30% of non-DS-AMKL cases and reported as a poor prognostic factor in pediatric AMKL [2]. In AML, patients with CBFA2T3::GLIS2 fusion have specific gene expression signatures that cluster them independently from other patients with non-DS-AMKL [3]. AML with CBFA2T3::GLIS2 fusion has recently been classified as AML with other rare recurring translocations in the International Consensus Classification of Myeloid Neoplasms and Acute Leukemias [4], and as AML with other defined genetic alterations in the 5th edition of the World Health Organization Classification of Haematolymphoid tumours [5].
We report the first Korean case of AMKL with CBFA2T3::GLIS2 fusion. The Institutional Review Board of Severance Hospital, Seoul, Korea, approved this study (4-2022-1294) and waived the need for informed consent.
A 20-month-old girl with no significant past medical history presented with a fever and elevated C-reactive protein levels and was hospitalized at a tertiary hospital in October 2021. The patient’s fever persisted in wax and wane patterns, and a workup for a fever of unknown origin was done. Diffuse osteolytic lesions in the whole axial skeleton, pelvic bone, proximal femurs, skull base, and facial bone were observed in computed tomography. The patient was transferred to the hemato-oncology division, and a bone marrow test was performed. The complete blood count was as follows: white blood cell (WBC) count 5.13×109/L; hemoglobin 9.4 g/dL; and platelet count 208×109/L. Blasts were 1 and 23.4% in peripheral blood and bone marrow (BM), respectively, negative in peroxidase and nonspecific esterase stains and positive in the periodic acid-Schiff stain. Flow cytometry analysis showed that the blasts were positive for CD33, CD117, CD41, CD34, and CD45 and negative for CD3, CD5, CD7, CD10, CD19, CD20, CD22, CD13, CD14, CD64, HLA-DR, cCD3, cCD22, cCD79a, cMPO, and TdT, indicating the presence of AMKL. Chromosome analysis showed the following karyotype: 46,XX,t(5,16)(q22;q23),inc[5]/46,XX[15] (Fig. 1A). Fluorescence in situ hybridization (FISH) was negative for CBFB::MYH11, RUNX1::RUNX1T1, PML::RARA, and KMT2A rearrangements. Multiplex reverse transcription PCR using a HemaVision kit (DNA Technology, Aarhus, Denmark) revealed no recurrent fusions. The patient received induction chemotherapy for three days. However, there was extravasation after chemoport insertion. The patient was referred to Severance Hospital for Hickmann catheterization and continued chemotherapy.
Subsequently, a second BM biopsy was performed. The BM contained blasts at 12.9% (Fig. 1B), and flow cytometry analysis revealed blasts with megakaryocytic differentiation and positive for CD33, CD117, CD41, CD61, and CD34 (Fig. 2). This was a rare case of AMKL that expressed CD34, as most AMKL cases are negative for CD34. No significant somatic variants were observed in a next-generation sequencing panel targeting 497 genes related to hematologic neoplasms. A targeted RNA fusion panel (FusionPlex Pan-Heme Panel; ArcherDX, CO, USA) detected 67 fusion reads between exon 9 of CBFA2T3 and exon 2 of GLIS2 (97.1% of total reads). The identified fusion transcript was confirmed using in-house RT-PCR and direct sequencing (Figs. 1C and 1D). A schematic representation of the CBFA2T3::GLIS2 fusion is provided in Fig. 1E.
Induction chemotherapy with cytarabine and idarubicin was administered for ten days. The patient suffered from Klebsiella pneumoniae sepsis, K. pneumoniae cellulitis, and Clostridium difficile infection during this time. The symptoms resolved, and cytarabine and mitoxantrone were administered for a second induction therapy. After one month of chemotherapy, the patient reached morphological complete remission (CR) with blasts reduced to 4.5%. Then only 0.9% and 2.9% of blasts were observed after two and four months of chemotherapy, respectively. Measurable residual disease (MRD) was assessed using flow cytometry and in-house RT-PCR. In the flow cytometry analysis, residual blasts were reported up to two months after chemotherapy and were not observed at four months. Because of these residual blasts, the regimen of the patient was changed to fludarabine, cytarabine, and idarubicin in the fourth month of chemotherapy. With the RT-PCR, the CBFA2T3::GLIS2 fusion transcript was detected up to four months after chemotherapy; therefore, molecular CR was not reached. The patient suffered neutropenic fever in the fifth month after treatment. K. pneumoniae was identified in the blood culture, and the patient expired from septic shock.
The CBFA2T3 gene is located at 16q24.3 and encodes a member of the myeloid translocation gene family, which interacts with transcription factors and functions as a transcriptional repressor via interaction with corepressor complexes [3]. The GLIS2 gene is located in 16p13.3 and encodes a nuclear transcription factor that has a role in Hedgehog pathway signaling [3]. CBFA2T3::GLIS2 fusion between two transcriptional regulators results in aberrant expression of the genes controlled by CBFA2T3 or GLIS2, which plays a role in megakaryoblastic leukemia development [6]. Patients with CBFA2T3::GLIS2 fusion usually have few somatic mutations and a poor prognosis [7, 8], as in our case. Overall survival for five years with CBFA2T3::GLIS2 is 22.0% versus 63.0% in fusion-negative cases [9]. Clinical data and test results between our case and previous reports of patients with non-Down syndrome pediatric AMKL in Korea were compared in Table 1.
There was a disparity between the chromosome result and the RNA fusion panel test. The t(5;16)(q22;q23) was the chromosome result obtained by another institution where the patient first visited. Due to poor chromosome quality, “inc” (i.e., an incomplete karyotype) was used in the nomenclature. No related fusion transcript was detected by the RNA fusion panel. The possible explanations are that t(5,16)(q22;q23) does not generate fusion transcripts or that the targeted RNA fusion panel does not include fusion genes of t(5,16)(q22;q23) in their targets. The t(5;16)(q22;q23) was not previously reported as related to hematologic malignancy. Therefore, the relationship of t(5;16)(q22;q23) with leukemogenesis cannot be ruled out. Detecting CBFA2T3::GLIS2 fusion using conventional cytogenetics, FISH, or commercial multiplex RT-PCR (e.g., the HemaVision test) is challenging. The inv(16)(p13.3q24.3), accompanied by CBFA2T3::GLIS2 fusion, is cytogenetically cryptic [2, 3]. Although FISH probes targeting CBFA2T3 and GLIS2 can detect the fusion, it is inefficient to use many probes to detect recurrent fusions observed in AML. Additionally, the HemaVision test can only detect 28 recurrent fusions and not CBFA2T3::GLIS2.
There was a significant difference between the percentage of fusion transcript reads (97.1% of total reads) and the bone marrow blast percentage (12.6% of all nucleated cells). Regarding RNA sequencing, reads are obtained from transcriptionally active regions [10]. Therefore, the percentage of the CBFA2T3::GLIS2 fusion transcript detected in RNA sequencing might differ from that of bone marrow blasts because normal blood cells have low CBFA2T3 and GLIS2 expressions.
In conclusion, we describe the first Korean report of non-DS-AMKL with CBFA2T3::GLIS2 fusion. Although a multiplex RT-PCR panel did not detect recurrent fusion, a targeted RNA fusion panel detected CBFA2T3::GLIS2 fusion. Additionally, in MRD assessed using in-house RT-PCR, CBFA2T3::GLIS2 fusion was observed to be more sensitive than flow cytometry. Thus, RNA fusion analysis could be a helpful tool to identify actionable genomic markers for patient risk stratification and MRD monitoring.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF-2021R1I1A1A01045980).
REFERENCES
1. Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. 1976; Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol. 33:451–8. DOI: 10.1111/j.1365-2141.1976.tb03563.x. PMID: 188440.
2. Masetti R, Pigazzi M, Togni M, Astolfi A, Indio V, Manara E, et al. 2013; CBFA2T3-GLIS2 fusion transcript is a novel common feature in pediatric, cytogenetically normal AML, not restricted to FAB M7 subtype. Blood. 121:3469–72. DOI: 10.1182/blood-2012-11-469825. PMID: 23407549.
3. Masetti R, Bertuccio SN, Pession A, Locatelli F. 2019; CBFA2T3-GLIS2-positive acute myeloid leukaemia. A peculiar paediatric entity. Br J Haematol. 184:337–47. DOI: 10.1111/bjh.15725. PMID: 30592296. PMCID: PMC6590351.
4. Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. 2022; International consensus classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood. 140:1200–28. DOI: 10.1182/blood.2022015850. PMID: 35767897.
5. Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. 2022; The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia. 36:1703–19. DOI: 10.1038/s41375-022-01613-1. PMID: 35732831. PMCID: PMC9252913.
6. Thiollier C, Lopez CK, Gerby B, Ignacimouttou C, Poglio S, Duffourd Y, et al. 2012; Characterization of novel genomic alterations and therapeutic approaches using acute megakaryoblastic leukemia xenograft models. J Exp Med. 209:2017–31. DOI: 10.1084/jem.20121343. PMID: 23045605. PMCID: PMC3478932.
7. Gruber TA, Larson Gedman A, Zhang J, Koss CS, Marada S, Ta HQ, et al. 2012; An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell. 22:683–97. DOI: 10.1016/j.ccr.2012.10.007. PMID: 23153540. PMCID: PMC3547667.
8. de Rooij JD, Branstetter C, Ma J, Li Y, Walsh MP, Cheng J, et al. 2017; Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet. 49:451–6. DOI: 10.1038/ng.3772. PMID: 28112737. PMCID: PMC5687824.
9. Smith JL, Ries RE, Hylkema T, Alonzo TA, Gerbing RB, Santaguida MT, et al. 2020; Comprehensive Transcriptome Profiling of cryptic CBFA2T3-GLIS2 fusion-positive AML defines novel therapeutic options: a COG and TARGET Pediatric AML study. Clin Cancer Res. 26:726–37. DOI: 10.1158/1078-0432.CCR-19-1800. PMID: 31719049. PMCID: PMC7002196.
10. Haas BJ, Dobin A, Li B, Stransky N, Pochet N, Regev A. 2019; Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 20:213. DOI: 10.1186/s13059-019-1842-9. PMID: 31639029. PMCID: PMC6802306.
11. Cho HJ, Park HK, Kong SY, Park Q, Kim SH, Ku HH. 2003; A case of acute megakaryoblastic leukemia diagnosed by t (1; 22)(p13; q13). Korean J Lab Med. 23:371–4.
12. Seo BY, Choi HW, Kang MG, Cho D, Kee SJ, Kim SH, et al. 2015; Constitutional chromosomal abnormality identified in a sibling donor after bone marrow stem cell transplantation in a pediatric patient with acute megakaryoblastic leukemia. Ann Lab Med. 35:162–4. DOI: 10.3343/alm.2015.35.1.162. PMID: 25553302. PMCID: PMC4272953.
Fig. 1
Morphological and genetic analysis. (A) G-banding karyotyping revealed 46,XX,t(5,16)(q22;q23),inc[5]/46,XX[15]. (B) Blasts on a bone marrow aspiration smear (Wright–Giemsa stain, 1,000×). (C) In-house reverse transcription (RT)-PCR using cDNA and (D) subsequent Sanger sequencing confirmed breakpoints in exons 9 and 2 of CBFA2T3 and GLIS2, respectively. (E) Schematic representation of the CBFA2T3::GLIS2 fusion.
Abbreviations: NTC, non-template control; NHR, nervy homology regions; MYND, myeloid, nervy, and DEAF-1; ex, exon; TAD, topologically associating domain; TRD, trans-repression domain; ZF, zinc finger.
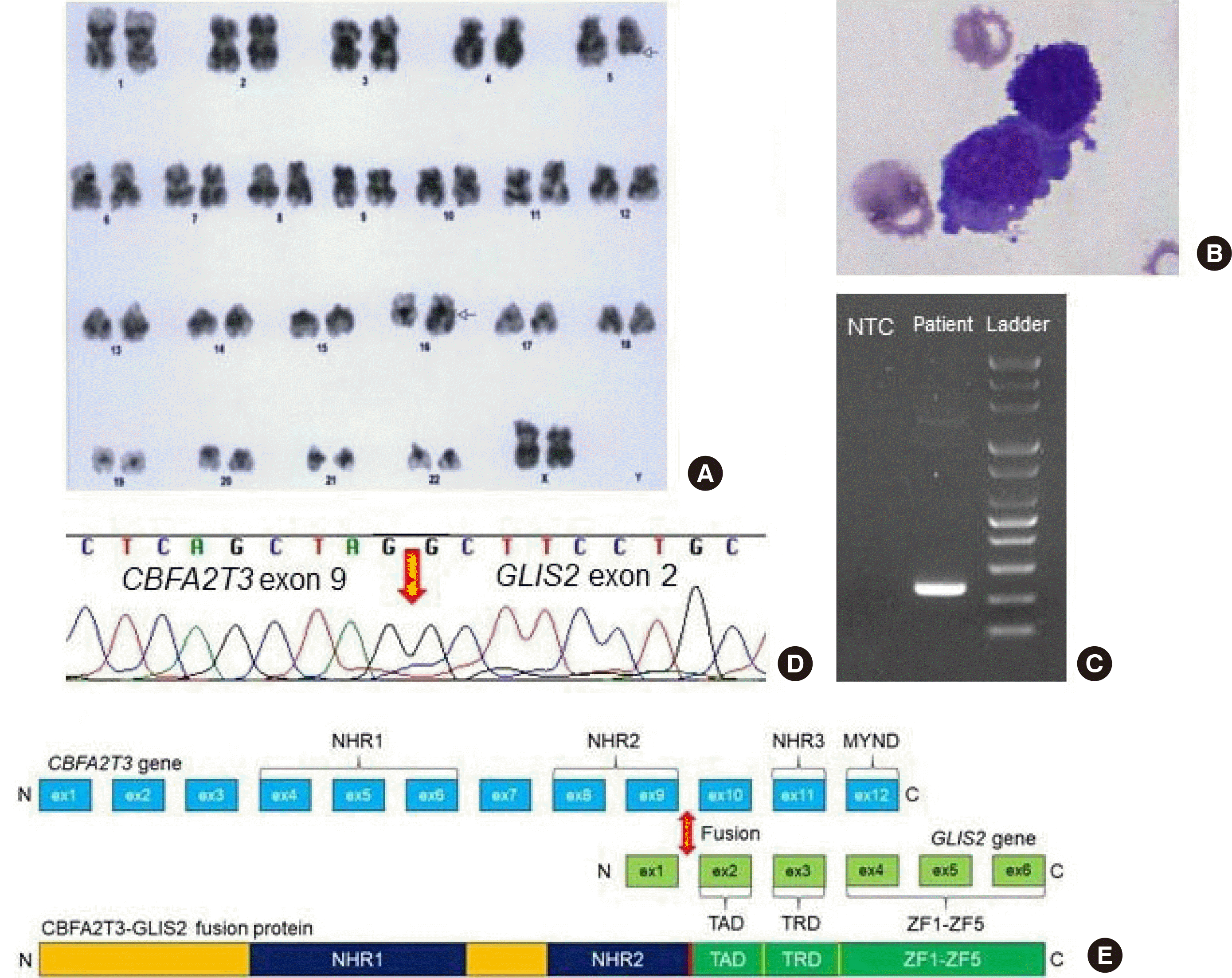
Fig. 2
Flow cytometry findings. Abnormal leukemic cells highlighted in blue (12.28% of total cells) exhibited a CD33+, CD117+, CD41+dim, CD61+dim, and CD34+ phenotype.
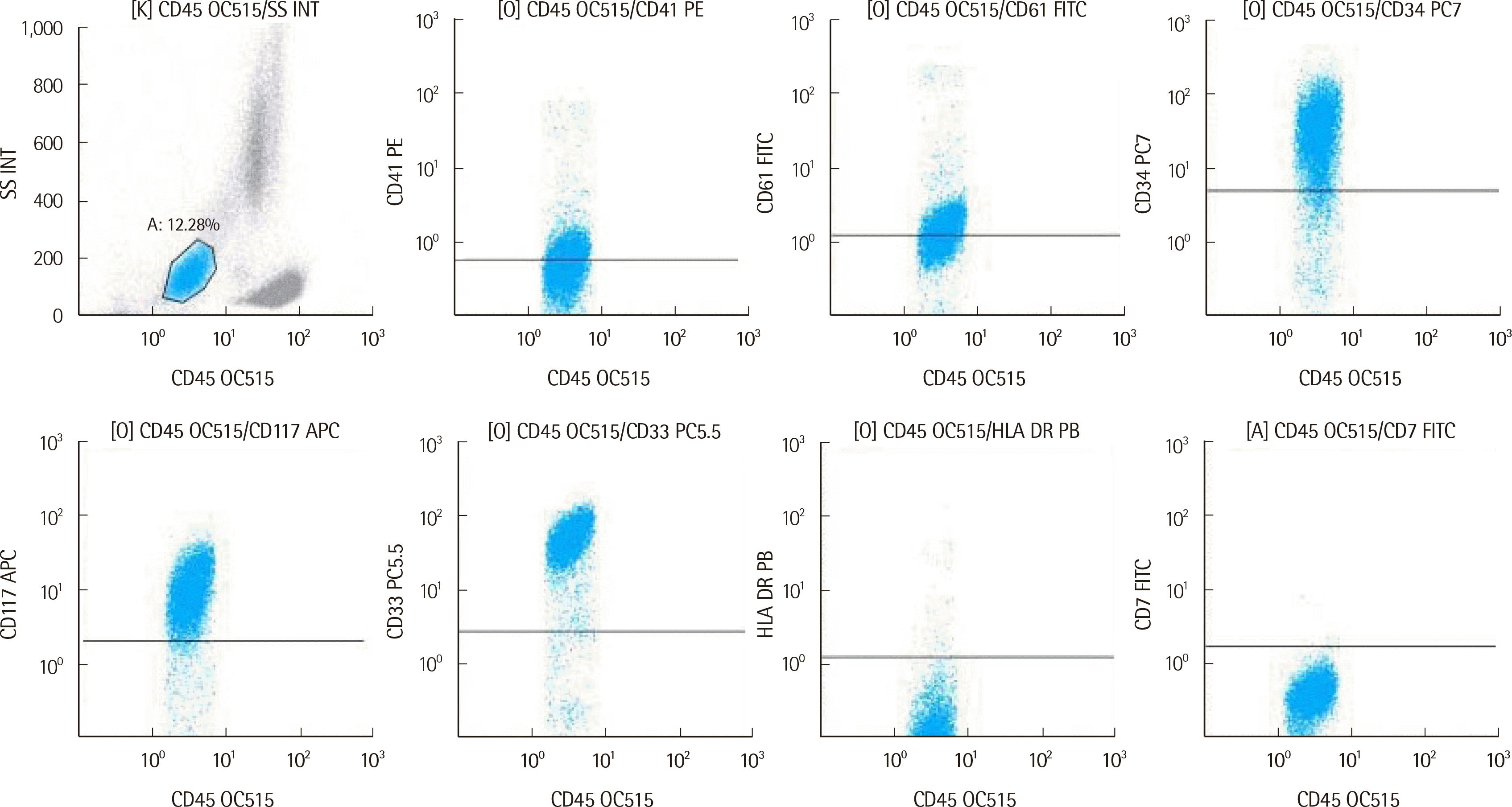
Table 1
Summary of reported non-Down syndromic pediatric acute megakaryocytic leukemia in Korea
| Characteristics | Present case | Cho, et al. [11] | Seo, et al. [12] |
|---|---|---|---|
| Sex | Female | Female | Male |
| Age | 1Y8M | 4Y | 10M |
| Complete blood count |
WBC count: 5.13×109/L with 1% blasts Hb: 9.4 g/dL Platelet count: 208×109/L |
WBC count: 4.630×109/L with a few blasts Hb: 8.4 g/dL Platelet count: 132×109/L |
WBC count: 46.41×109/L |
| Bone marrow | 23.4% of blasts with irregular cytoplasmic blebs | Dry-tapped marrow | 68% of blasts with irregular cytoplasmic blebs |
| Flow cytometry | Positive for CD33, CD117, CD41, CD34, and CD45 | Positive for CD13, CD33, CD34, HLA-DR, and CD61 | Positive for CD41 and HLA-DR |
| Chromosome | 46,XX,t(5,16)(q22;q23),inc[5]/46,XX[15] | 48,XX,t(1;22)(p13;q13),+der(1) t(1;22),+2[1]/46,XX [19] | 45,XY,-22,t(2;11)(q32;q23),der(14)t(14;22)[6]/46,XY,t(2;11)(q32;q23)[8] |
| Gene rearrangement | CBFA2T3::GLIS2 fusion | Not tested | Not tested |
| Somatic mutation | None | Not tested | Not tested |
| Treatment and outcome | Died from septic shock five months after diagnosis | Complete remission for 8 months after induc- tion chemotherapy using enocitabine, idarubi- cin, cytarabin, and thiguanine followed by consolidation chemotherapy using low-dose cytosine | Complete remission after allogenic bone marrow transplantation |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download