Abstract
Background
Immune responses to infectious disease differ according to disease severity. In this study, we investigated the overall laboratory profiles, including antibody titers and cytokine levels in unvaccinated patients with Coronavirus Disease 2019 (COVID-19), according to disease severity at the initial stage during the early pandemic period.
Methods
In total, 195 unvaccinated patients with COVID-19, who were hospitalized from February to August 2020 were enrolled in this study. The patients were divided into five severity categories, and their antibody titers and cytokine levels were evaluated by collecting serial blood samples up to the first three months after infection.
Results
Older male patients with comorbidities had a higher probability of rapid disease progression. Patients who required intensive care showed significantly lower initial hemoglobin, platelet counts, total protein, and albumin levels, with significantly higher neutrophil-to-lymphocyte ratios, blood urea nitrogen, creatinine, and coagulation, cardiac, and inflammatory markers compared to those in patients who did not require intensive care. At three weeks after infection, the titers of both the anti-receptor binding domain of spike protein and neutralizing antibodies were significantly higher in patients with high disease severity than in patients with low disease severity; a similar trend was observed for IL-6 and TNF-α levels for at least three months from infection onset.
초록
배경
감염병 환자의 면역반응은 질병의 중증도에 따라 다르게 나타난다. 본 연구에서는 코로나19 팬데믹 초기의 백신 투여력이 없는 코로나19 환자들을 중증도에 따라 분류하고, 항체 역가와 사이토카인 수치를 포함한 전반적인 검사실 혈액 검사 결과를 조사하였다.
방법
2020년 2월부터 8월까지 백신 투여력이 없는 195명의 코로나19 환자를 모집하였다. 각 환자는 질환의 중증도에 따라 5개의 카테고리로 분류하였고, 감염 후 최대 3개월까지 채혈하여 항체 역가와 사이토카인 수치를 파악하였다.
An infectious disease involves an external agent, a host, and the environment. Therefore, hosts infected with the same pathogen (agent) in the same environment may show variations in disease progression and prognosis depending on the host characteristics [1, 2]. The novel coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [3, 4]. The Centers for Disease Control and Prevention estimated that over 57% of the US population and 75% US children have been infected by SARS-CoV-2 until February 2022 [3]. Further, the World Health Organization has announced that COVID-19 has resulted in 616,427,419 confirmed cases and 6,528,557 deaths as of October 5, 2022 [4]. The clinical manifestations of SARS-CoV-2 infection exhibit a wide range of severity, from asymptomatic and mild to severe pneumonia, myocarditis, acute kidney injury, respiratory distress syndrome, multiorgan failure, and death. Various studies have reported that the severity of COVID-19 symptoms is related to the host epidemiological characteristics, hematological and immunological profile, similar to that in other infectious diseases [5-13]. In previous studies, the disease severity and mortality rates have been reported to differ depending on the study period. The laboratory profiles of infected patients showed increased white blood cell counts, neutrophil-lymphocyte ratios, troponin, D-dimer, and c-reactive protein levels, and decreased platelet counts and hemoglobin [5-8]. Unfortunately, early studies in the pandemic were limited in terms of complete investigation, including varying severities, and laboratory test parameters, while the latest studies had analytical bias owing to the influence of vaccine inoculation and various SARS-CoV-2 variants.
The composition and function of the human immune response vary among individuals and are influenced by several factors [13-15], including specific antibodies (immunoglobulins) to the pathogen. Antibodies represent humoral immunity and are generated in response to external stimuli such as pathogens. Therefore, the presence or absence of antibodies can be used to evaluate whether an individual has been previously infected with a pathogen [16, 17]. Further, antibodies have protective and therapeutic effects and are commonly used as therapeutic agents. COVID-19 mRNA vaccines, the most widely used COVID-19 vaccine type, use a specific mechanism to activate antibody production [18-20]. Monoclonal antibody therapies, such as casirivimab/imdevimab (REGEN-COV™, Regeneron Pharmaceuticals, Inc., Tarrytown, NY, USA), regdanvimab (Regkirona™, Celltrion, Inc., Incheon, Korea), sotrovimab (Xevudy™, GlaxoSmithKline LLC, Zebulon, NC, USA), and tixagevimab/cilgavimab (Evusheld™, AstraZeneca, Cambridge, UK) have also been actively administered to high-risk patients with COVID-19 worldwide [21-23].
Cytokines are important components of the human immune system and are secreted by various immune cells such as macrophages, lymphocytes, and mast cells to send signals to other cells. They are classified into several families according to their structures. The major cytokine families include the interleukins (ILs), members of the tumor necrosis factor (TNF) superfamily, interferons, and chemokines family. Each family has dozens to hundreds of cytokines, each of which promotes or suppresses inflammation and regulates cellular functions [24, 25]. Cytokine kinetics change dynamically over the course of disease and play important roles in the response to common viral infections [26]. Therefore, IL-6, IL-8, IL-1β, TNF-α, and interferon-γ which show potential as anti-inflammatory therapies, are being actively studied in patients with COVID-19 [11-13, 27, 28].
In this study, we recruited and collected samples from patients diagnosed with COVID-19 (strongly presumed to be the wild-type, Wuhan strain) in the early pandemic period, before the emergence of major SARS-CoV-2 variants and vaccine development, to exclude variant and vaccine effects. Furthermore, we performed a broader and more in-depth investigation than that in previous studies on the relationship between patient baseline characteristics, serial antibody titers, serial cytokine (IL-6 and TNF-α) levels, and COVID-19 severity for up to three months after SARS-CoV-2 infection.
Patients diagnosed with COVID-19 (based on a reverse-transcription polymerase chain reaction test of a nasopharyngeal or oropharyngeal sample) and hospitalized at Boramae Medical Center, a designated Korean COVID-19 treatment center from February to August 2020, were included in the study. None of the patients had been vaccinated against COVID-19, and all participants were thought to have been infected with wild-type SARS-CoV-2. The time (days) from infection was calculated based on the date of symptom onset in symptomatic patients and the date of diagnosis in asymptomatic patients. The days from infection were divided into five intervals: 0–7 days (1 week), 8–14 days (2 weeks), 15–21 days (3 weeks), 22–42 days (4–6 weeks), and 43–84 days (7–12 weeks).
The study was approved by the Institutional Review Board of Boramae Medical Center (IRB 21-2021-38); consent was waived because the study involved no more than minimal risk. Moreover, the waiver did not adversely affect the rights and welfare of the participants, and the research could not have been feasibly conducted without the waiver being granted. Whenever appropriate, the participants would be provided with additional pertinent information after participation.
The clinical severity of SARS-CoV-2 infection was classified from asymptomatic infection to critical illness according to the clinical features and treatment required based on the established guidelines [29-31]. Patients without symptoms were classified as having “asymptomatic infection”; those with mild symptoms, which could be controlled through only supportive care, and without dyspnea were classified as having “mild illness”; patients with clinical and radiographic evidence of lower respiratory tract disease, who were administered oxygen via nasal prongs or a face mask were classified as having “moderate illness”; those with oxygen saturation <94%, representing severe pneumonia, or treated with specific therapies such as dexamethasone, were classified as having “severe illness”; and patients with respiratory failure, shock, or multiorgan dysfunction or failure were classified as having “critical illness”.
The main bases of outcomes were requiring oxygen treatment (“asymptomatic and mild” group or “moderate, severe, and critical” group) and requiring intensive care unit care (“asymptomatic, mild and moderate” group or “severe and critical” group).
In addition to collection of history, the patient underwent physical examination, chest X-ray, and initial blood sampling within two days of hospital admission. The blood tests included the complete blood counts (hemoglobin, white blood cells, and platelets), a chemistry panel (blood urea nitrogen, creatinine, protein, albumin, bilirubin, alkaline phosphatase, aspartate transaminase, and alanine aminotransferase), a coagulation panel (prothrombin time, activated partial thromboplastin time, fibrinogen, and D-dimer), cardiac markers (creatine kinase, creatine kinase-myocardial band, troponin I, and pro-B-type natriuretic peptide), and inflammatory markers (C-reactive protein, procalcitonin, lactate dehydrogenase, and ferritin).
Blood collection was performed using a serum separator tube followed by serum separation via centrifugation at 3,000 rpm for 10 minutes. The samples were then stored at -80°C until use. All tests were performed within one month of blood collection.
Anti-nucleocapsid (N) antibody: An automated electrochemiluminescence immunoassay (ECLIA)-based Elecsys Anti-SARS-CoV-2 assay (Elecsys Anti-N; Roche Diagnostics, Mannheim, Germany) was performed using a Cobas 800/e 801 analyzer (Roche Diagnostics). The Elecsys Anti-N assay uses a recombinant protein representing the nucleocapsid (N) antigen to qualitatively detect anti-SARS-CoV-2 antibodies. The results were recorded as reactive (positive) or non-reactive (negative) depending on whether the cutoff index was ≥1.0 or <1.0, according to the manufacturer’s directions.
Anti-spike/receptor-binding domain (S/RBD) antibody: An automated ECLIA-based quantitative Elecsys Anti-SARS-CoV-2 S assay (Elecsys Anti-S/RBD; Roche Diagnostics) was performed using a Cobas 800/e 801 analyzer. The Elecsys Anti-S/RBD assay uses a recombinant protein representing the receptor binding domain (RBD) of the spike (S) antigen, which enables quantitative detection of high-affinity anti-SARS-CoV-2 antibodies. Titers ≥0.8 U/mL were reported as reactive (positive), whereas those <0.8 U/mL were reported as non-reactive (negative) according to the manufacturer’s instructions.
Neutralizing antibody: To identify neutralizing antibodies, we used the cPass SARS-CoV-2 Neutralization Antibody Detection kit ver. RUO 3.0, (cPass Neutralizing Antibody; GenScript Biotech, Piscataway, NJ, USA), an enzyme-linked immunosorbent assay (ELISA)-based surrogate virus neutralization test (sVNT). The results are based on the inhibition rate. According to the manufacturer’s instructions, the cut-off value is 30%; therefore, if neutralizing antibodies were detected to be ≥30%, the result was considered as positive, whereas values <30% were considered as negative. This cut-off value is based on validation of sera from patients with COVID-19 and healthy controls, and is reported to have 100.0% (95% CI: 87.1–100.0%) and 100.0% (95% CI: 95.8–100.0%) positive percent agreement with the plaque reduction neutralization tests, PRNT50 and PRNT90, respectively.
IL-6: An automated ECLIA-based Elecsys IL-6 (Roche Diagnostics) was performed using a Cobas 800/e 801 analyzer. According to the manufacturer’s instructions, the reference value is ≤7 pg/mL.
TNF-α: The Quantikine® HS ELISA (R&D Systems, Minneapolis, MN, USA) is a 4 h solid phase ELISA designed to measure human TNF-α in serum. This assay employs the quantitative sandwich enzyme immunoassay technique. According to the manufacturer’s instructions, the reference value is ≤10 pg/mL.
Data were compared using the independent samples t-test or Mann-Whitney U-test for continuous variables and the chi-square test for categorical variables. Pearson correlation analysis was used to evaluate the relationship between the Elecsys Anti-S/RBD and cPass Neutralizing Antibody results. Two-sided P values of P<0.05 were considered statistically significant. Multivariate analysis (logistic regression analysis) was performed to eliminate the influence of confounding variables and explain the degree of association among various risk factors. Selected correlates with P<0.2 in the univariable analysis were included in the multivariable logistic regression model. All statistical analyses were performed using SPSS version 27.0 (SPSS, Inc., Chicago, IL, USA) and Excel software (Microsoft Corporation, Redmond, WA, USA).
In total, 195 patients were included in the study, from whom 274 serum samples were collected within 84 days of infection. The number of samples obtained in each time interval was as follows: 96 in days 0–7, 72 in days 8–14, 34 in days 15–21, 53 in days 22–42, and 19 in days 43–84. The samples were obtained in only one interval from 137 patients, two intervals from 42 patients, three intervals from 12 patients, four intervals from three patients, and all five intervals from only one patient.
The demographic and clinical characteristics of the 195 participants are shown in Table 1. Forty-five patients (23.1%) were less than 40 years of age, 85 (43.6%) were between 40–64 years of age, and 65 (33.3%) were aged ≥65 years. There were 94 female patients (48.2%) and 101 male patients (51.8%). Sixty-nine patients (35.4%) were previously healthy, and 126 (64.6%) had underlying conditions such as hypertension, diabetes mellitus, coronary artery disease, chronic lung disease, or cancer. Most participants (96.4%) were Koreans. Of the patients, 139 (71.3%) reported general symptoms, 125 (64.1%) reported respiratory symptoms, 22 (11.3%) reported loss of taste and smell, and 18 (9.2%) reported gastrointestinal symptoms. Further, 153 patients (78.5%) had asymptomatic or mild illness, 23 (11.8%) had moderate illness, and 19 (9.7%) had severe or critical illness.
Of the 156 patients who could specify the date of symptom onset, an average of 3.6, 4.3, and 5.4 days passed from symptom onset to diagnosis, hospital admission, and radiography and first blood sampling, respectively. Further, during the analysis period, the quarantine release criteria of South Korean domestic COVID-19 guidelines were eased (as of June 25, 2020), after which the average hospital stay of patients who were asymptomatic and had mild illness was significantly reduced from 23.7 to 17.4 days (P=0.002).
The disease severity and outcome according to patient age, sex, and underlying disease status are shown in Fig. 1. The probability of an unfavorable outcome increased significantly with increasing age and the presence of comorbidities; further, the probability of requiring ventilator/intensive care unit care was significantly higher in males than in females.
The relation between laboratory data at the initial blood sampling and COVID-19 severity is shown in Table 2. In the severe and critical (high severity) group who later required intensive care, the initial hemoglobin, platelet, total protein, and albumin levels were significantly lower and the neutrophil-to-lymphocyte ratio, blood urea nitrogen, and creatinine levels were significantly higher than those in the asymptomatic, mild, and moderate (low severity) group. In addition, coagulation, inflammatory, and cardiac markers were significantly higher in the high severity group.
Most patients had anti-SARs-CoV-2 antibodies starting at two weeks (15 days) from the date of infection and maintained these antibodies for up to three months. The antibody-positive rates were somewhat higher in patients with low severity illness before two weeks and were somewhat higher in patients with high severity illness after two weeks, but these differences were not significant (Fig. 2).
The anti-S/RBD and neutralizing antibody titers were significantly higher in patients with high severity disease than in patients with low severity disease at the time of initial antibody production at approximately three weeks after infection (Fig. 3A, B). Cytokine (IL-6 and TNF-α) levels were consistently higher in patients with high severity disease than in patients with low severity disease from immediately after infection until at least three months (Fig. 3C, D).
The neutralizing antibodies and the anti-S/RBD antibodies in the Elecsys Anti-SARS-CoV-2 S assay and cPass SARS-CoV-2 Neutralization Antibody Detection Kit (ELISA-based sVNT method) showed a significantly positive correlation (r=0.540, P<0.001). The cPass Neutralizing Antibody Kit cut-off value of 30.0%, suggested by GenScript, was equivalent to approximately 340 U/mL using the Elecsys Anti-SARS-CoV-2 S assay (data not shown).
The parameters included in the multivariable logistic regression analysis were age, sex, underlying disease, and initial X-ray findings. The results of logistic regression analysis are shown in Table 3.
During the initial period of the SARS-CoV-2 epidemic, South Korea responded with very strict quarantine restrictions and detailed investigations and disclosures regarding the movement of all people known to be infected. According to this national policy, all people with respiratory symptoms, close contacts with infected patients, and foreign visitors entering the country were required to undergo a PCR test, and those with confirmed infection were required to be admitted to a medical facility as soon as infection was confirmed. Because this study was performed at the beginning of the epidemic, all patients were hospitalized regardless of symptoms; thus, patients were not selected according to disease severity. Additionally, all study participants were interviewed about their medical history and underwent a timely SARS-CoV-2 PCR test, blood collection, and radiographic imaging. After October 2020, the national policy was changed, and home quarantine and non-face-to-face treatment was implemented; thus, detailed data on patients with asymptomatic and mild illness could not be collected. Therefore, research on the early stages of the pandemic is important for analyzing the full spectrum of COVID-19 clinical manifestations.
Several previous studies have shown that cancer, cerebrovascular disease, chronic kidney disease, liver cirrhosis, diabetes mellitus, disabilities, and cardiovascular disease are high risk factors for severe outcomes in COVID-19 [8, 32, 33]; however, we found no significant associations between underlying disease and COVID-19 severity. This may be because the underlying disease was identified based on the patient questionnaire, and the comorbidities were well-controlled at the time of admission. To reduce the potential for bias related to underlying disease, it is necessary to collect objective data and obtain accurate data on the patient condition at the time of hospitalization.
As in previous studies [9-11], serum antibodies against SARS-CoV-2 were detected by the third week (15 days) of infection in all study participants, and the patients showing high severity illness had high titers of S/RBD antibodies and neutralizing antibodies. Furthermore, there was no significant difference in the antibody titer in the first two weeks of infection (before 14 days), but this difference was significant in the third week.
In agreement with previous studies [11-13, 34-38], the pro-inflammatory cytokines, IL-6 and TNF-α, showed higher levels in patients with more severe disease than in those with less severe disease. Therefore, inhibitors of pro-inflammatory cytokines may be useful as therapies for reducing disease severity. Although, differences in infectivity and cytokine secretion were observed among variants such as alpha (B.1.1.7), beta (B.1.351), delta (B.1.617.2), and omicron (B.1.1.529) [39], a cytokine pattern similar to that of the wild-type strain was observed as indicated by several studies during the specific variant outbreak period showing similar results [34-38]. Based on these cytokine dynamics, several new drugs targeting IL-6 and TNF-α have been developed to prevent cytokine storms [12]. These drugs have various mechanisms of action, including blocking cytokine receptors, promoting anti-cytokine antibody production, and reducing cytokine levels to suppress their function, and have shown promising results [12, 40-42].
Overall, this study clearly highlights the immunological characteristics of unvaccinated patients with confirmed SARS-CoV-2 infections in South Korea. However, it does have some limitations. First, it has a retrospective, single-center design with a small sample size. As the center is located in Seoul, most patients were residents of Seoul, and the possibility of regional bias could not be ruled out. Second, serial samples through consecutive three-month intervals were only obtained from a few patients, which makes it difficult to examine individual antibody titers and cytokine level dynamics.
We showed that the levels of anti-S/RBD and neutralizing antibodies as well as of pro-inflammatory cytokines, IL-6 and TNF-α, varied significantly according to disease severity during the early stage of SARS-CoV-2 infection. Although we examined the early stages of the COVID-19 pandemic, individuals with more recent SARS-CoV-2 infection and those with other respiratory viruses may exhibit similar trends. Therefore, the immunological characteristics of patients confirmed through blood sampling at the appropriate time may be useful for predicting clinical outcomes to provide effective treatments for patients with respiratory viral infections. Further, we focused on infections with the early wild-type SARS-CoV-2, providing comparative data for studying the differences among variant types in further studies. COVID-19 has greatly impacted society as well as led to research and clinical data collection, which can advance medical technology.
Acknowledgements
This work was supported by the Seoul National University Hospital Research Fund [grant no 04-2018-0610]. The reagent kits, Elecsys® Anti-SARS-CoV-2 and Elecsys® Anti-SARS-CoV-2 S were provided by Roche Diagnostics.
REFERENCES
1. Rothman KJ. 1976; Causes. Am J Epidemiol. 104:587–92. DOI: 10.1093/oxfordjournals.aje.a112335. PMID: 998606.
2. Center for Disease Control and Prevention. Concepts of Disease Occurrence. https://www.cdc.gov/csels/dsepd/ss1978/lesson1/section8.html. Updated Nov 2011.
3. Clarke KEN, Jones JM, Deng Y, Nycz E, Lee A, Iachan R, et al. 2022; Seroprevalence of infection-induced SARS-CoV-2 antibodies - United States, September 2021-February 2022. MMWR Morb Mortal Wkly Rep. 71:606–8. DOI: 10.15585/mmwr.mm7117e3. PMID: 35482574. PMCID: PMC9098232.
4. World Health Organization. Coronavirus (COVID-19) Dashboard. https://covid19.who.int/. Last accessed on Oct 6th 2022.
5. Kim SW, Kim SM, Kim YK, Kim JY, Lee YM, Kim BO, et al. 2021; Clinical characteristics and outcomes of COVID-19 cohort patients in Daegu metropolitan city outbreak in 2020. J Korean Med Sci. 36:e12. DOI: 10.3346/jkms.2021.36.e12. PMID: 33398946. PMCID: PMC7781854.
6. Surme S, Buyukyazgan A, Bayramlar OF, Cinar AK, Copur B, Zerdali E, et al. 2021; Predictors of intensive care unit admission or mortality in patients with Coronavirus disease 2019 pneumonia in Istanbul, Turkey. Jpn J Infect Dis. 74:458–64. DOI: 10.7883/yoken.JJID.2020.1065. PMID: 33642427.
7. Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. 2020; Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 58:1021–8. DOI: 10.1515/cclm-2020-0369. PMID: 32286245.
8. Gallo Marin B, Aghagoli G, Lavine K, Yang L, Siff EJ, Chiang SS, et al. 2021; Predictors of COVID-19 severity: a literature review. Rev Med Virol. 31:1–10. DOI: 10.1002/rmv.2146. PMID: 32845042. PMCID: PMC7855377.
9. Dispinseri S, Secchi M, Pirillo MF, Tolazzi M, Borghi M, Brigatti C, et al. 2021; Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat Commun. 12:2670. DOI: 10.1038/s41467-021-22958-8. PMID: 33976165. PMCID: PMC8113594.
10. Maeda T, Kashiwagi K, Yoshizawa S, Sato T, Aoki K, Ishii Y, et al. 2021; Early anti-SARS-CoV-2 immunoglobulin G response may be associated with disease severity in patients with COVID-19. Jpn J Infect Dis. 74:560–2. DOI: 10.7883/yoken.JJID.2020.799. PMID: 33642431.
11. Kwon JS, Kim JY, Kim MC, Park SY, Kim BN, Bae S, et al. 2020; Factors of severity in patients with COVID-19: cytokine/chemokine concentrations, viral load, and antibody responses. Am J Trop Med Hyg. 103:2412–8. DOI: 10.4269/ajtmh.20-1110. PMID: 33124544. PMCID: PMC7695090.
12. Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L. 2020; SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 54:62–75. DOI: 10.1016/j.cytogfr.2020.06.001. PMID: 32513566. PMCID: PMC7265853.
13. Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. 2020; Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front Immunol. 11:1949. DOI: 10.3389/fimmu.2020.01949. PMID: 32849654. PMCID: PMC7426442.
14. Brodin P, Davis MM. 2017; Human immune system variation. Nat Rev Immunol. 17:21–9. DOI: 10.1038/nri.2016.125. PMID: 27916977. PMCID: PMC5328245.
15. Marshall JS, Warrington R, Watson W, Kim HL. 2018; An introduction to immunology and immunopathology. Allergy Asthma Clin Immunol. 14(Suppl 2):49. DOI: 10.1186/s13223-018-0278-1. PMID: 30263032. PMCID: PMC6156898.
16. Cooper NR, Nemerow GR. 1984; The role of antibody and complement in the control of viral infections. J Invest Dermatol. 83:121s–7s. DOI: 10.1111/1523-1747.ep12281847. PMID: 6376646.
17. Brady LJ. 2005; Antibody-mediated immunomodulation: a strategy to improve host responses against microbial antigens. Infect Immun. 73:671–8. DOI: 10.1128/IAI.73.2.671-678.2005. PMID: 15664904. PMCID: PMC547018.
18. Jalkanen P, Kolehmainen P, Häkkinen HK, Huttunen M, Tähtinen PA, Lundberg R, et al. 2021; COVID-19 mRNA vaccine induced antibody responses against three SARS-CoV-2 variants. Nat Commun. 12:3991. DOI: 10.1038/s41467-021-24285-4. PMID: 34183681. PMCID: PMC8239026.
19. Doria-Rose N, Suthar MS, Makowski M, O'Connell S, McDermott AB, Flach B, et al. 2021; Antibody persistence through 6 months after the second dose of mRNA-1273 vaccine for Covid-19. N Engl J Med. 384:2259–61. DOI: 10.1056/NEJMc2103916. PMID: 33822494. PMCID: PMC8524784.
20. Kim N, Minn D, Park S, Roh EY, Yoon JH, Park H, et al. 2021; Positivity of SARS-CoV-2 antibodies among Korean healthy healthcare workers 1 and 2 weeks after second dose of Pfizer-BioNTech vaccination. J Korean Med Sci. 36:e158. DOI: 10.3346/jkms.2021.36.e158. PMID: 34060264. PMCID: PMC8167405.
21. Yang L, Liu W, Yu X, Wu M, Reichert JM, Ho M. 2020; COVID-19 antibody therapeutics tracker: a global online database of antibody therapeutics for the prevention and treatment of COVID-19. Antib Ther. 3:205–12. DOI: 10.1093/abt/tbaa020. PMID: 33215063. PMCID: PMC7454247.
22. Deb P, Molla MMA, Saif-Ur-Rahman KM. 2021; An update to monoclonal antibody as therapeutic option against COVID-19. Biosaf Health. 3:87–91. DOI: 10.1016/j.bsheal.2021.02.001. PMID: 33585808. PMCID: PMC7872849.
23. Zheng J, Deng Y, Zhao Z, Mao B, Lu M, Lin Y, et al. 2022; Characterization of SARS-CoV-2-specific humoral immunity and its potential applications and therapeutic prospects. Cell Mol Immunol. 19:150–7. DOI: 10.1038/s41423-021-00774-w. PMID: 34645940. PMCID: PMC8513558.
24. Dinarello CA. 2007; Historical insights into cytokines. Eur J Immunol. 37(Suppl 1):S34–45. DOI: 10.1002/eji.200737772. PMID: 17972343. PMCID: PMC3140102.
25. Zhang J-M, An J. 2007; Cytokines, inflammation, and pain. Int Anesthesiol Clin. 45:27–37. DOI: 10.1097/AIA.0b013e318034194e. PMID: 17426506. PMCID: PMC2785020.
26. Beltra JC, Decaluwe H. 2016; Cytokines and persistent viral infections. Cytokine. 82:4–15. DOI: 10.1016/j.cyto.2016.02.006. PMID: 26907634.
27. Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. 2020; The COVID-19 cytokine storm; what we know so far. Front Immunol. 11:1146. DOI: 10.3389/fimmu.2020.01446. PMID: 32612617. PMCID: PMC7308649.
28. Del Valle DM, Kim-Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, et al. 2020; An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 26:1636–43. DOI: 10.1038/s41591-020-1051-9. PMID: 32839624. PMCID: PMC7869028.
29. National Institutes of Health. Clinical Spectrum of SARS-CoV-2 Infection. https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/. Last updated on Sep 2022.
30. Gandhi RT, Lynch JB, Del Rio C. 2020; Mild or moderate Covid-19. N Engl J Med. 383:1757–66. DOI: 10.1056/NEJMcp2009249. PMID: 32329974.
31. Korea Disease Control and Prevention Agency. 2022. Coronavirus (COVID-19) guideline, Republic of Korea (local government). 13rd-1 ed. Central Disaster Management Headquarters and Central Disease Control Headquarters.
32. Center for Disease Control and Prevention. Underlying medical conditions associated with higher risk for severe COVID-19: Information for healthcare professionals. https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-care/underlyingconditions.html. Last accessed on Feb 2022.
33. Sanyaolu A, Okorie C, Marinkovic A, Patidar R, Younis K, Desai P, et al. 2020; Comorbidity and its impact on patients with COVID-19. SN Compr Clin Med. 2:1069–76. DOI: 10.1007/s42399-020-00363-4. PMID: 32838147. PMCID: PMC7314621.
34. Basheer M, Saad E, Assy N. 2022; The cytokine storm in COVID-19: the strongest link to morbidity and mortality in the current epidemic. COVID. 2:540–52. DOI: 10.3390/covid2050040.
35. Niedźwiedzka-Rystwej P, Majchrzak A, Kurkowska S, Małkowska P, Sierawska O, Hrynkiewicz R, et al. 2022; Immune signature of COVID-19: In-depth reasons and consequences of the cytokine storm. Int J Mol Sci. 23:4545. DOI: 10.3390/ijms23094545. PMID: 35562935. PMCID: PMC9105989.
36. Jing X, Xu M, Song D, Yue T, Wang Y, Zhang P, et al. 2022; Association between inflammatory cytokines and anti-SARS-CoV-2 antibodies in hospitalized patients with COVID-19. Immun Ageing. 19:12. DOI: 10.1186/s12979-022-00271-2. PMID: 35248063. PMCID: PMC8897556.
37. Schultheiß C, Willscher E, Paschold L, Gottschick C, Klee B, Henkes SS, et al. 2022; The IL-1β, IL-6, and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep Med. 3:100663. DOI: 10.1016/j.xcrm.2022.100663. PMID: 35732153. PMCID: PMC9214726.
38. Ling L, Chen Z, Lui G, Wong CK, Wong WT, Ng RWY, et al. 2021; Longitudinal cytokine profile in patients with mild to critical COVID-19. Front Immunol. 12:763292. DOI: 10.3389/fimmu.2021.763292. PMID: 34938289. PMCID: PMC8685399.
39. Do TND, Claes S, Schols D, Neyts J, Jochmans D. 2022; SARS-CoV-2 virion infectivity and cytokine production in primary human airway epithelial cells. Viruses. 14:951. DOI: 10.3390/v14050951. PMID: 35632693. PMCID: PMC9144593.
40. Nyberg T, Ferguson NM, Nash SG, Webster HH, Flaxman S, Andrews N, et al. 2022; Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. 399:1303–12. DOI: 10.1016/S0140-6736(22)00462-7. PMID: 35305296.
41. Servellita V, Syed AM, Morris MK, Brazer N, Saldhi P, Garcia-Knight M, et al. 2022; Neutralizing immunity in vaccine breakthrough infections from the SARS-CoV-2 Omicron and Delta variants. Cell. 185:1539–48.e5. DOI: 10.1016/j.cell.2022.03.019. PMID: 35429436. PMCID: PMC8930394.
42. Korea Disease Control and Prevention Agency. 2021; July 2021 status and characteristics of the COVID-19 variant virus outbreak in the Republic of Korea. Public Health Wkly Rep. 14:3388–96.
Fig. 1
Effects of age, sex, and underlying disease on the prognosis of patients with COVID-19. The upper bar graph shows the distribution of disease severity according to age [young (≤40 years), middle (41–64 years), old (≥65 years)], sex (female, male), and underlying disease (present or absent). The bottom table shows the odds ratios for COVID-19 severity by age, sex, and underlying disease. The odds ratios were calculated based on two criteria, one for requiring oxygen treatment (left) and one for requiring intensive care unit care (right).
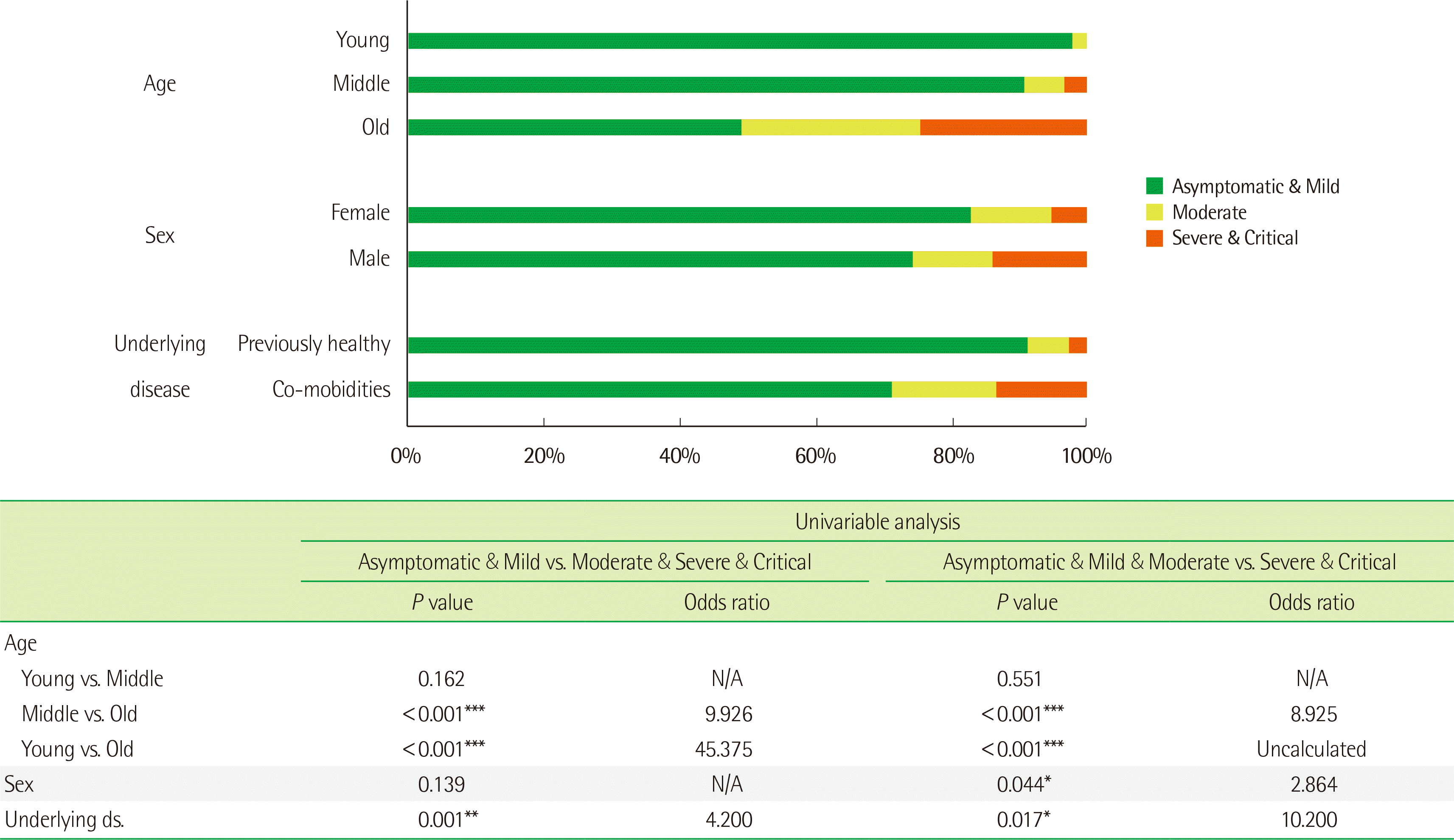
Fig. 2
SARS-CoV-2 antibody positivity rates in the early stage of infection according to disease severity. The days from infection were divided into five intervals, and the (A) anti-nucleocapsid (anti-N), (B) anti-spike/receptor binding domain (anti-S/RBD), and (C) neutralizing antibody positivity rate between patients with asymptomatic & mild (low severity) illness and patients with moderate and severe & critical (high severity) illness were compared. The number of samples obtained in each time interval was as follows: 96 in days 0–7 (73 low severity and 23 high severity), 72 in days 8–14 (46 low severity and 26 high severity), 34 in days 15–21 (28 low severity and 6 high severity), 53 in days 22–42 (43 low severity and 10 high severity), and 19 in days 43–84 (15 low severity and 4 high severity).
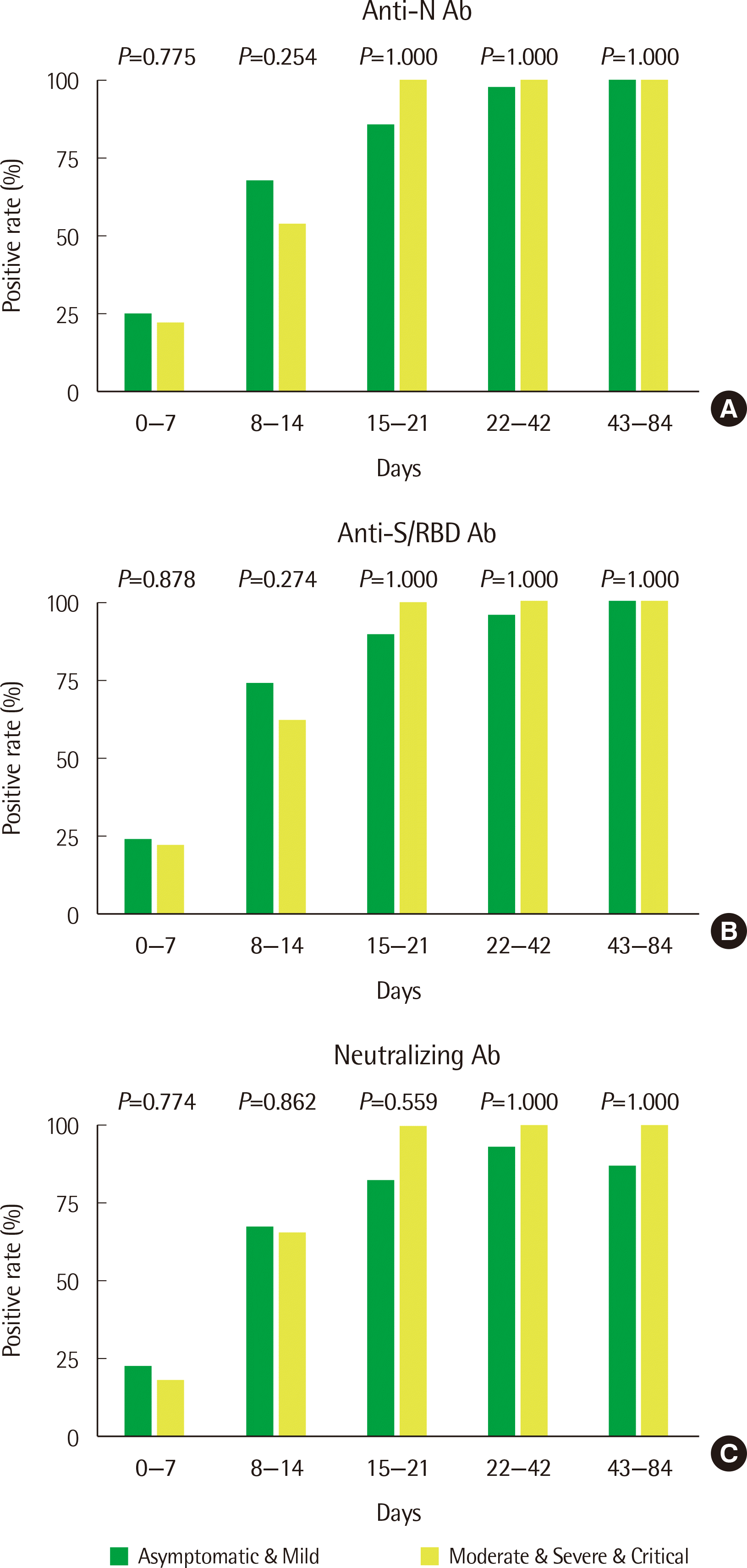
Fig. 3
Box-and-whiskers plots comparing antibody titers and cytokine levels according to disease severity. The days from infection were divided into five intervals, and the (A) anti-spike/receptor binding domain (anti-S/RBD) antibody titer, (B) neutralizing antibody titer, (C) IL-6 level, and (D) TNF-α level were compared between patients with asymptomatic and mild (low severity) illness and patients with moderate, severe, and critical (high severity) illness. The number of samples obtained in each time interval was as follows: 96 in days 0–7 (73 low severity and 23 high severity), 72 in days 8–14 (46 low severity and 26 high severity), 34 in days 15–21 (28 low severity and 6 high severity), 53 in days 22–42 (43 low severity and 10 high severity), and 19 in days 43–84 (15 low severity and 4 high severity). The box-and-whiskers plot comprises a box showing the interquartile range, line showing the median, cross showing the mean, whiskers showing the range, and marks showing the outlying values.
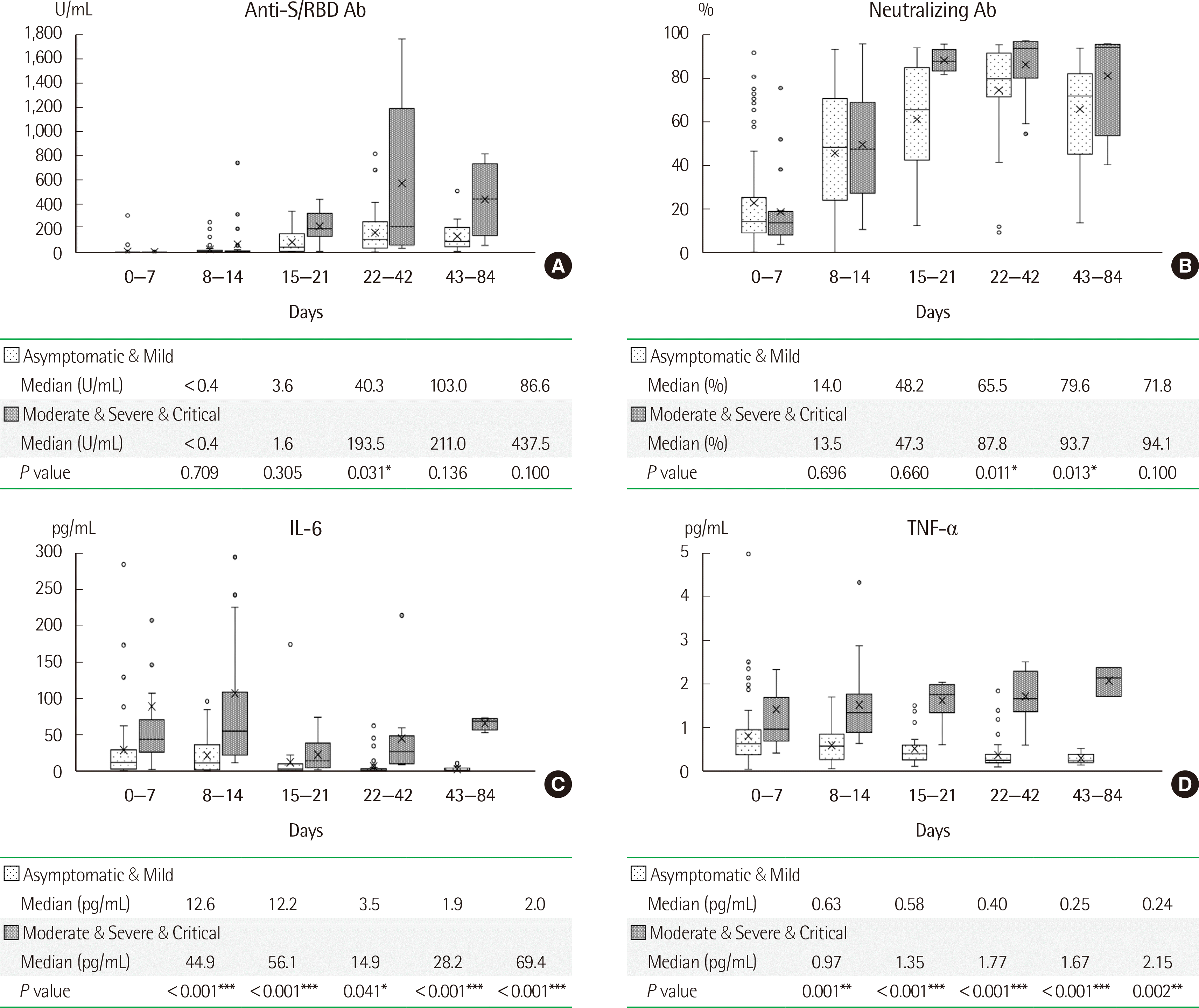
Table 1
Baseline characteristics of the 195 study participants
| Characteristics | Enrolled subjects (N=195) |
|---|---|
| Demographic | N (%) |
| Age group | |
| Young < 40 yr | 45 (23.1) |
| Middle 40–64 yr | 85 (43.6) |
| Old ≥ 65 yr | 65 (33.3) |
| Sex | |
| Female | 94 (48.2) |
| Male | 101 (51.8) |
| Underlying disease | |
| Previously healthy | 69 (35.4) |
| Co-morbidities (HTN, DM, coronary artery disease, chronic lung disease, cancer etc.) | 126 (64.6) |
| Ethnicity and race | |
| Korean | 188 (96.4) |
| Other Asian (non-Korean) | 3 (1.5) |
| American and European | 4 (2.1) |
| Disease severity classification | N (%) |
| Asymptomatic & mild | 153 (78.5) |
| Moderate | 23 (11.8) |
| Severe & critical | 19 (9.7) |
| Symptoms at diagnosis | N (%) |
| General symptom (fever, fatigue, myalgia, headache) | 139 (71.3) |
| Respiratory symptom (cough, sputum, sore throat, runny nose, dyspnea) | 125 (64.1) |
| Loss of taste/smell | 22 (11.3) |
| Gastrointestinal symptom (nausea, vomiting, diarrhea) | 18 (9.2) |
| Mean days from symptom onset | days (95% CI) |
| To diagnosis confirmed (N = 156) | 3.6 (2.8–4.4) |
| To hospital admission (N = 156) | 4.3 (3.5–5.1) |
| To check chest X-ray and blood sampling at BRMH (N = 156) | 5.4 (4.2–6.6) |
| Mean days of hospital stay | |
| Total patients | |
| Before 25th June, 2020* (N = 69) | 24.4 (20.6–28.3) |
| After 25th June, 2020* (N = 122) | 22.3 (19.5–25.2) |
| Asymptomatic & mild symptom patients | |
| Before 25th June, 2020* (N = 67) | 23.7 (20.1–27.3) |
| After 25th June, 2020* (N = 86) | 17.4 (16.2–18.7) |
Table 2
Comparison of initial laboratory data according to clinical severity
Values are presented as median (interquartile rage Q1–Q3). Data were compared using the Mann-Whitney U test.
Abbreviations: Hb, hemoglobin; WBC, white blood cell; NLR, neutrophil-to-lymphocyte ratio; BUN, blood urea nitrogen; ALP, alkaline phosphatase; AST, aspartate transaminase; ALT, alanine aminotransferase; PT, prothrombin time; APTT, activated partial thromboplastin time; CK, creatine kinase; CK-MB, creatine kinase-myocardial band; Pro-BNP, pro-B-type natriuretic peptide; CRP, C-reactive protein.
Table 3
Multivariable logistic regression analysis of demographic and clinical parameters contributing to intensive care unit care of COVID-19




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download