Abstract
Background
Liquid chromatography/tandem mass spectrometry (LC-MS/MS)-based assays are increasingly being used for therapeutic drug monitoring (TDM) of immunosuppressive drugs (ISDs). However, many laboratories that employ LC-MS/MS assays for TDM of ISDs use manual sample preparation steps.
Methods
We implemented a semi-automated sample preparation method based on protein precipitation, using an Andrew+ liquid handler, integrated with an LC-MS/MS-based TDM assay for four ISDs (tacrolimus, sirolimus, everolimus, and cyclosporine A). The feasibility and analytical performance were analyzed and compared with those of the manual sample preparation method using 163 clinical samples.
Results
The total execution time of Andrew+-based sample preparation method was 55 minutes (including off-line centrifugation) for 48 samples. However, the hands-on time was significantly reduced than that of the manual sample preparation method. The average repeatability and within-laboratory precision of the assay were acceptable (CV ≤10%), and the results of manual and Andrew+-based sample preparation methods were equivalent.
Conclusions
The sample preparation method using the Andrew+ liquid handler was successfully integrated with LC-MS/MS-based TDM assays for ISDs. Sample preparation using Andrew+ was reproducible, standardized, and enabled excellent utilization of technical staff time. Although further optimization for full automation is still pursued, this study indicates the potential of integrating liquid handers with LC-MS/MS-based assays in the clinical setting.
초록
배경
면역억제제의 치료적 약물농도 감시를 위한 액체크로마토그래피-탠덤질량분석기의 도입은 임상검사실에서 그 활용이 점차 늘어나고 있다. 그러나 대부분의 임상검사실에서 치료적 약물농도 감시를 위한 질량분석법을 위해 반드시 필요한 검체 전처리 과정은 대부분 수작업으로 이루어지고 있다.
방법
본 연구에서 저자들은 Andrew+ liquid handler를 활용하여 단백질 침전의 원리를 적용하는 반자동 검체 전처리 방법을 개발하고 이를 질량분석법에 접목시킨 4종 면역억제제(tacrolimus, sirolimus, everolimus, 그리고 cyclosporine A)의 치료적 약물농도 감시 검사를 평가하였다. 즉, 반자동 검체 전처리 방법의 유용성을 평가하고 총 163개의 임상 검체를 이용하여 검사 수행능을 분석하였으며 이를 수작업 검체 전처리법과 비교 평가하였다.
Therapeutic drug monitoring (TDM) of immunosuppressive drugs (ISDs) optimizes their efficacy and minimizes toxicity in patients with organ transplantation and other autoimmune disorders [1]. The rationale behind this recommendation includes a narrow therapeutic index, large inter- and intra-individual pharmacokinetic variability, and severe consequences of both ISD overexposure and underexposure. For this purpose, clinical laboratories providing TDM of ISDs primarily utilize immunoassays (such as enzyme-multiplied, affinity chrome-mediated, chemiluminescent microparticle, and electrochemiluminescence immunoassays) or liquid chromatography/tandem mass spectrometry (LC-MS/MS). The analytical performance of LC-MS/MS-based assays is superior to that of immunoassays in terms of analytical accuracy and specificity [2], and LC-MS/MS is considered a reference method [3]. However, the labor-intensive and time-consuming sample preparation is a major drawback of using LC-MS/MS methods for TDM of ISDs.
Meanwhile, liquid handlers have been successfully used for the automation of workflows in clinical laboratories, including LC-MS/MS automation [4]. While most advancements in LC-MS/MS automation have originated from the pharmaceutical industry, the need for automation in medical diagnostic application of LC-MS/MS is nonetheless compelling. Hence, this study aimed to apply and optimize a liquid handler platform, Andrew+ (Waters, Milford, MA, USA) to the sample preparation workflow (based on protein precipitation) for LC-MS/MS-based TDM of four ISDs (tacrolimus, sirolimus, everolimus, and cyclosporine A) and analyze the relevant analytical performances related to the sample preparation method.
All patient samples used in this study were residual samples collected for routine LC-MS/MS-based ISD assays. Whole blood samples were collected in Vacuette vacuum tubes (Greiner Bio-One GmbH, Kremsmünster, Austria) containing EDTA as anticoagulant. This study was reviewed and approved by the Institutional Review Board of Seoul St. Mary’s Hospital (KC22DISE0794), and a waiver of informed consent was granted.
Non-filtered, color-coded Optifit Tips compatible with Andrew+ were obtained from Sartorius (Goettingen, Germany). Ninety-six-well sample collection plates with 2 mL square wells, corresponding adhesive seals, and 96-well sample collection plates with 350 µL round wells were obtained from Waters. A six-column reagent reservoir was obtained from Agilent (Santa Clara, CA, USA).
LC-MS-grade methanol and formic acid were purchased from Honeywell Burdick & Jackson (Muskegon, MI, USA). Ammonium acetate and zinc sulfate heptahydrate were obtained from Sigma-Aldrich. Deionized water was generated using a Millipore Milli-Q Gradient Water Purification System (Molsheim, France). The 6PLUS1 multilevel whole blood calibrator sets and MassCheck whole blood controls were obtained from Chromsystems (Munich, Germany). The protein precipitation solution was 0.1 M ZnSO4. The internal standard working solution contained 10 ng/mL ascomycin (Sigma-Aldrich, Burlington, MA, USA) and 10 µg/mL cyclosporin D (United States Biological, Salem, MA, USA) in acetonitrile.
The liquid handling platform, Andrew+ pipetting robot (Waters), consists of a tool arm that can hold and charge up to four pipettes and a robotic arm for pipette and plate handling. Andrew+ also uses labware holders called dominos, and up to two full rows of dominos can be installed. In addition, by adding Device+ (Waters), processes such as heating, shaking, vacuum application, or labware transportation can be incorporated. The Andrew+ pipetting robot is controlled using the OneLab+ software (Waters) [5].
For the ISD assay, two multichannel Andrew Alliance Picus Pipettes (120 and 1,200 µL, Waters), a single-channel Andrew Alliance Picus Pipette (120 µL, Waters), two tip insertion system dominos, two conical microtube dominos, a deep well microplate domino, a microplate domino, and a microplate shaker+ device (Waters) were utilized (Fig. 1). The tip insertion system dominos were used to hold disposable pipette tips; the conical microtube dominos (for up to 24 samples each) were used for calibrators, controls, and samples; the deep well microplate domino was used for the six-column reagent reservoir that contained the protein precipitation and internal standard solutions; the microplate domino was used for the injection plate; and the microplate shaker+ device was used for the sample collection plate (reaction plate). An Eppendorf Centrifuge 5810R (Eppendorf SE, Hamburg, Germany) was used for off-line centrifugation.
The ISD assay workflow using Andrew+ consisted of three basic steps: sample transfer, protein precipitation, and sample analysis. A few manual steps were required, including the initial experimental setup, off-line centrifugation, and loading the injection plate onto the autosampler. The workflow process is described in detail in Fig. 2. The workflow and related pipetting parameters were designed and translated into OneLab executable scripts using an intuitive interface provided by the OneLab+ software. For off-line centrifugation, the plate was sealed and briefly mixed on a microplate shaker+ device and then centrifuged at 13,400× g for 10 minutes on an Eppendorf Centrifuge 5810R.
An ACQUITY UPLC I-Class System (Waters) equipped with a Binary Solvent Manager and flow-through needle Sample Manager was used for chromatographic separation. For chromatographic separation, 20 µL of extracted sample was injected onto an ACQUITY UPLC HSS C18 SB column (1×100 mm, 1.8 µm particle size, 55.0°C column temperature, Waters). The mobile phase consisted of solution A (0.1% formic acid and 2 mM ammonium acetate in deionized water) and solution B (0.1% formic acid and 2 mM ammonium acetate in methanol) with the following linear gradient: initial, 50% B; 0.2–0.6 minutes, 50% B–100% B; 0.6–1.2 minutes, 100% B–50% B. The total run time was 1.8 minutes, with 0.4 mL/min constant flow rate.
UPLC was coupled to a Xevo™ TQD IVD MS (Waters). The electrospray ionization source was operated in the positive mode with the following parameters: capillary voltage, 3 kV; cone voltage, 34 V; source temperature, 150°C; desolvation temperature, 400°C; nitrogen desolvation gas flow, 1000 L/h; and nitrogen cone gas flow, 10 L/h. The compounds were detected via multiple reaction monitoring mode employing the following ion transitions: m/z 809.70 → 756.50 (collision energy: 22 V) for ascomycin, m/z 821.50 → 768.50 (collision energy: 22 V) for tacrolimus, m/z 931.60 → 864.50 (collision energy: 16 V) for sirolimus, m/z 975.60 → 908.60 (collision energy: 18 V) for everolimus, m/z 1,219.80 → 1,202.80 (collision energy: 22 V) for cyclosporine A, and m/z 1,233.95 → 1,216.80 (collision energy: 22 V) for cyclosporin D. The data were processed using the MassLynx™ Software v4.2 (Waters).
The total process time, hands-on time, feasibility of integration with the LC-MS/MS-based TDM of ISDs, and analytical performance of Andrew+-based UPLC-MS/MS method were evaluated. Three levels of MassCheck whole blood-based quality controls (ChromSystems) were utilized for precision evaluation. In addition, 163 samples (45 tacrolimus, 37 sirolimus, 36 everolimus, and 44 cyclosporine A), encompassing the target range of respective ISDs (i.e., tacrolimus 5–20 ng/mL, sirolimus 5–15 ng/mL, everolimus 3–8 ng/mL, cyclosporine A 150–300 ng/mL), were analyzed using both the manual and Andrew+-based sample preparation methods, and the results were compared. Analyse-it for Microsoft Excel (Analyse-it Software, Ltd., Leeds, UK) was used for data processing. Passing–Bablok nonparametric linear regression was used for method comparison.
The total execution time of the sample preparation method developed using Andrew+ pipetting system for quantification of four ISDs was 55 minutes (including 20 minutes for off-line centrifugation) for 48 samples (Table 1). Since a significant amount of time was allotted for off-line centrifugation (10 and 20 minutes for the manual and Andrew+-based preparation methods, respectively), it is safe to say that the total time required for preparing 48 samples was comparable between the manual and Andrew+ preparation methods. Meanwhile, the hands-on time was significantly reduced from 27 to 2 minutes when using the Andrew+ pipetting system.
The repeatability and within-laboratory precision of the assay assessed at three separate QC levels are shown in Table 2. Repeatability and within-laboratory precision were both acceptable and fulfilled the recommended performance criteria for imprecision, where applicable (CV ≤10% and ≤5% for 50 and 300 ng/mL cyclosporine A, respectively) [6].
The equations of the Passing–Bablok regression lines for all four ISDs are shown in Table 3. The estimates of parameters (slope and intercept with their lower and upper limits of the 95% confidence intervals) using the Passing–Bablok regression between the manual and Andrew+ sample preparation methods are shown in Table 2. For all four ISDs, the 95% confidence intervals of the slope and intercept encompassed 1 and 0, respectively; hence, the statistical differences between the results of the manual and Andrew+-based sample preparation methods integrated with LC-MS/MS were considered insignificant. Moreover, the bias and 95% CI of bias estimate, calculated from the regression line at relevant medical decision points of each ISDs (i.e., tacrolimus 5 ng/mL, sirolimus 5 ng/mL, everolimus 3 ng/mL, cyclosporine A 150 ng/mL) were within the prespecified acceptance criteria of 10%.
LC-MS/MS method is increasingly used for TDM of ISDs in clinical laboratories, mainly because of its high analytical specificity and multiplexing capability [7]. Published sample preparation methods for LC-MS/MS-based ISD assays include protein precipitation, liquid–liquid extraction [8], and online solid-phase extraction [9]. However, in many laboratories, the samples for LC-MS/MS assays for ISDs are prepared manually [10]. The Andrew+ pipetting robot, a versatile liquid handling system for increasing automation in the sample preparation steps for LC-MS/MS assays, has been recently introduced. While an application note from Andrew+ for semi-automated sample preparation for LC-MS/MS-based ISDs is available [11], to the best of our knowledge, this is the first report of using the Andrew+ pipetting system for LC-MS/MS-based TDM assays for ISDs using clinical samples. In this report, substituting manual extraction by the semi-automated protein precipitation method using the Andrew+ pipetting system allowed reproducible and standardized sample preparation workflow with less hands-on time for whole blood samples and provided comparable results for LC-MS/MS-based ISD quantification.
One of the challenges in adopting a liquid handler for sample preparation for TDM of ISDs is maintaining homogeneity of the whole blood sample, since the ISDs are partitioned into erythrocytes and the cellular components sediment within minutes when the sample is at rest. To ensure whole blood homogeneity, we applied resuspension procedures using repeated aspiration/dispensing, and the results were comparable to those of the manual preparation method, which utilizes mixing by agitation of primary blood collection tubes. In addition, we modified the off-line centrifugation force of the OneLab protocol of 2,000× g to 13,400× g. The configuration of the pipetting parameters for each liquid transfer was also optimized to avoid cross-well contamination and obtain the best achievable precision.
Equivalence in analytical performance (such as precision and comparison) was observed using Andrew+ for transferring samples, calibrators, QCs, and reagents, as well as adding internal standard and protein precipitation solutions and transferring extracted supernatants. However, there is still room for improvement, and further optimization could be pursued to achieve the full potential of automated sample preparation using the Andrew+ platform. For example, off-line centrifugation is a major non-automated step in the present workflow, necessitating a significant amount of time (20 minutes) and additional hands-on time. Alternatives to centrifugation after protein precipitation that can increase the degree of automation include vacuum filtration, which is a viable solution for full automation, particularly when a new vacuum pump, extraction+device, controlled via the OneLab Software, have been introduced. Another feature that could be optimized is the option to use primary blood collection tubes, with perhaps a primary tube cap piercing feature that could replace manual de-capping and sample transfer.
In summary, the results demonstrate the feasibility of integrating the Andrew+ liquid handler with LC-MS/MS-based TDM assays for ISDs. Sample preparation based on the protein precipitation method using Andrew+ is reproducible, provides standardized workflow, and significantly reduces hands-on time, which is one of the key drivers of adoption of automated sample preparation methods for LC-MS/MS. Moreover, the assay performance was equivalent to that of the manual protein extraction method. Although further optimization is being pursued, this study indicates the potential of integrating relatively simple liquid handlers with LC-MS/MS-based assays in the clinical laboratory.
Acknowledgments
This work was supported by the Korean Society of Clinical Chemistry (KSCC) Research Fund 2022(2). The authors also thank Yunung Kim for his involvement in the evaluation and Chiyeol Park for his management support.
REFERENCES
1. Seger C, Shipkova M, Christians U, Billaud EM, Wang P, Holt DW, et al. 2016; Assuring the proper analytical performance of measurement procedures for immunosuppressive drug concentrations in clinical practice: recommendations of the international association of therapeutic drug monitoring and clinical toxicology immunosuppressive drug scientific committee. Ther Drug Monit. 38:170–89. DOI: 10.1097/FTD.0000000000000269. PMID: 26982493.
2. Keevil BG, Tierney DP, Cooper DP, Morris MR. 2002; Rapid liquid chromatography-tandem mass spectrometry method for routine analysis of cyclosporin A over an extended concentration range. Clin Chem. 48:69–76. DOI: 10.1093/clinchem/48.1.69. PMID: 11751540.
3. Oellerich M, Armstrong VW, Schütz E, Shaw LM. 1998; Therapeutic drug monitoring of cyclosporine and tacrolimus. Clin Biochem. 31:309–16. DOI: 10.1016/S0009-9120(98)00049-6. PMID: 9721427.
4. Vogeser M, Kirchhoff F. 2011; Progress in automation of LC-MS in laboratory medicine. Clin Biochem. 44:4–13. DOI: 10.1016/j.clinbiochem.2010.06.005. PMID: 20599877.
5. Zucchelli P, Horak G, Skinner N. 2021; Highly versatile cloud-based automation solution for the remote design and execution of experiment protocols during the COVID-19 pandemic. SLAS Technol. 26:127–39. DOI: 10.1177/2472630320971218. PMID: 33210978. PMCID: PMC7684276.
6. Oellerich M, Armstrong VW, Kahan B, Shaw L, Holt DW, Yatscoff R, et al. 1995; Lake Louise Consensus Conference on cyclosporin monitoring in organ transplantation: report of the consensus panel. Ther Drug Monit. 17:642–54. DOI: 10.1097/00007691-199512000-00017. PMID: 8588235.
7. Kim HK, Park HD, Lee SG, Chae H, Song SH, Lee YW, et al. 2021; Immunosuppressive drug measurement by liquid chromatography coupled to tandem mass spectrometry: interlaboratory comparison in the Korean clinical laboratories. Ann Lab Med. 41:268–76. DOI: 10.3343/alm.2021.41.3.268. PMID: 33303711. PMCID: PMC7748092.
8. Kvamsøe MM, Hansen KR, Skadberg Ø, Vethe NT, Brede C. 2020; Salting out-assisted liquid-liquid extraction for liquid chromatography-tandem mass spectrometry measurement of tacrolimus, sirolimus, everolimus, and cyclosporine a in whole blood. Ther Drug Monit. 42:695–701. DOI: 10.1097/FTD.0000000000000794. PMID: 32701594.
9. Ceglarek U, Lembcke J, Fiedler GM, Werner M, Witzigmann H, Hauss JP, et al. 2004; Rapid simultaneous quantification of immunosuppressants in transplant patients by turbulent flow chromatography combined with tandem mass spectrometry. Clin Chim Acta. 346:181–90. DOI: 10.1016/j.cccn.2004.03.017. PMID: 15256319.
10. Deslandes G, Grégoire M, Renaud C, Monteil-Ganière C, Azoulay C, Pineau A, et al. 2016; Comparison between an automated and manual extraction for the determination of immunosuppressive drugs whole blood concentrations by liquid chromatography tandem mass spectrometry. J Clin Lab Anal. 30:924–9. DOI: 10.1002/jcla.21958. PMID: 27086934. PMCID: PMC6807118.
11. OneLab Design & Execute - Andrew Alliance. Immunosuppressant Sample Prep. https://onelab.andrewalliance.com/library/immunosuppressant-sample-prep-BE07g0RP. Last accessed on Jan 2023.
Fig. 1
Andrew+ pipetting robot system layout. 1) A tool arm that holds and charges up to four pipettes, 2) a robotic arm for pipette and plate handling, 3) two pipette tip insertion system dominos, 4) two conical microtube dominos, 5) a deep well microplate domino, 6) a microplate domino, and 7) a microplate shaker device.
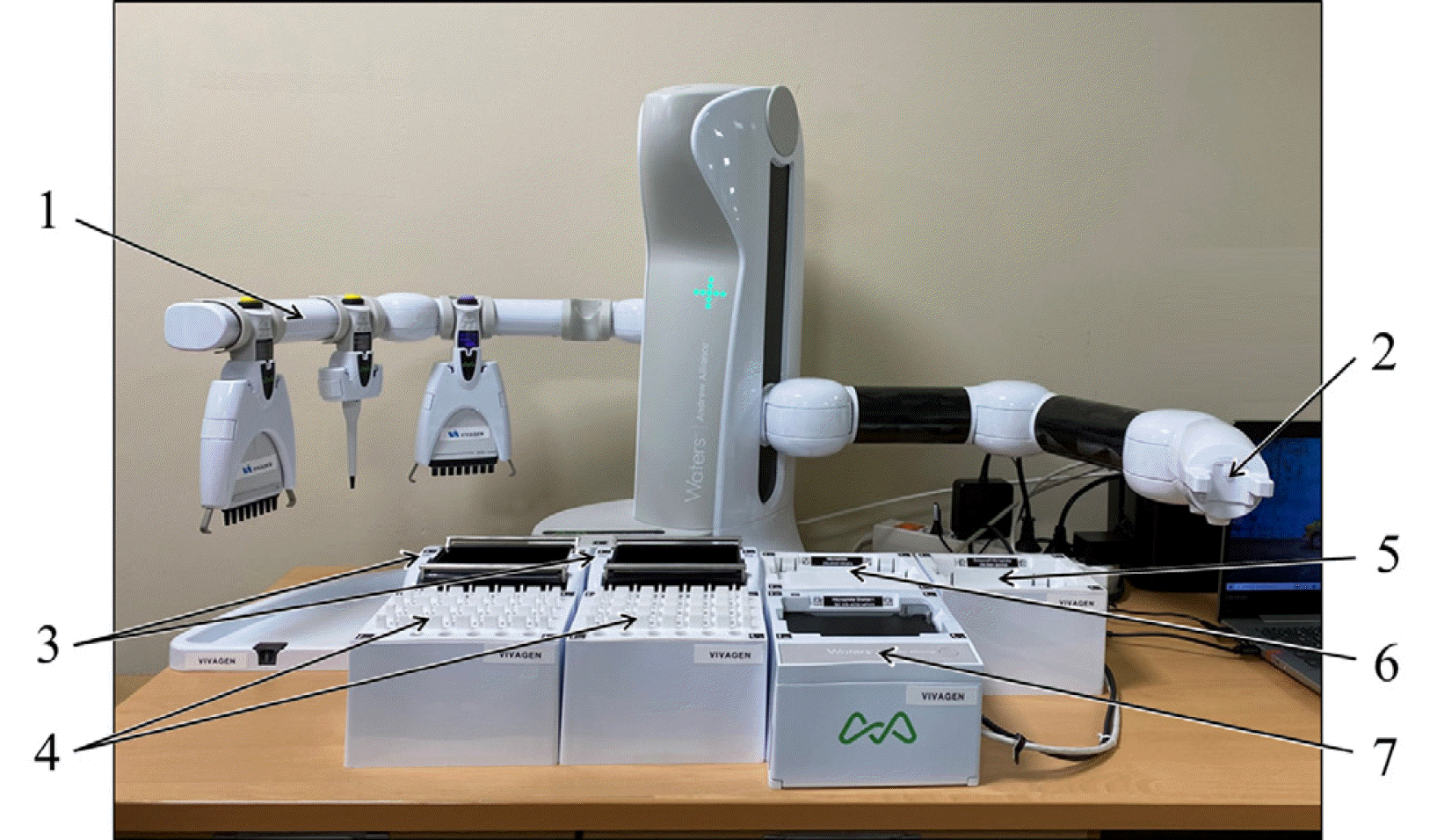
Fig. 2
Workflow of the sample preparation process using the Andrew+ pipetting robot system configured for liquid chromatography/tandem mass spectrometry immunosuppressant analysis.

Table 1
Comparison of total and hands-on times using manual vs. automated sample preparation methods used in this study for 48 samples
Table 2
Analytical imprecision of the Andrew+-based liquid chromatography/tandem mass spectrometry assay
| Analyte | QC Level | Mean (ng/mL) | Repeatability* | Within-laboratory Precision† | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| SD (ng/mL) | CV% | SD (ng/mL) | CV% | |||
| Tacrolimus | Level 1 | 2.6 | 0.18 | 7.1 | 0.24 | 9.2 |
| Level 2 | 7.0 | 0.31 | 4.4 | 0.41 | 5.8 | |
| Level 3 | 14.8 | 0.51 | 3.5 | 0.59 | 4.0 | |
| Sirolimus | Level 1 | 2.8 | 0.26 | 9.6 | 0.27 | 10.0 |
| Level 2 | 10.1 | 0.66 | 6.5 | 0.93 | 9.2 | |
| Level 3 | 19.3 | 1.06 | 5.5 | 1.40 | 7.2 | |
| Everolimus | Level 1 | 2.5 | 0.22 | 8.8 | 0.28 | 11.1 |
| Level 2 | 4.7 | 0.39 | 8.3 | 0.43 | 9.2 | |
| Level 3 | 9.0 | 0.41 | 4.6 | 0.55 | 6.1 | |
| Cyclosporine A | Level 1 | 46.9 | 1.48 | 3.1 | 2.45 | 5.2 |
| Level 2 | 248.3 | 4.24 | 1.7 | 5.37 | 2.2 | |
| Level 3 | 492.5 | 8.91 | 1.8 | 11.48 | 2.3 | |
Table 3
The estimates of Passing–Bablok regression between the manual (x) and Andrew+-based sample preparation (y) methods
| Analyte | Estimate | 95% CI | |
|---|---|---|---|
| Tacrolimus (n = 45) | Intercept | -0.6621 | -1.493 to 0.2000 |
| Slope | 1.103 | 1.000 to 1.213 | |
| Spearman’s ρ* | 0.953 | 0.913 to 0.974 | |
| Sirolimus (n = 37) | Intercept | 0.2000 | -0.8571 to 1.183 |
| Slope | 1.0000 | 0.8280 to 1.149 | |
| Spearman’s ρ | 0.952 | 0.907 to 0.976 | |
| Everolimus (n = 36) | Intercept | 0.1778 | 0 to 0.5392 |
| Slope | 0.9751 | 0.8824 to 1.032 | |
| Spearman’s ρ | 0.975 | 0.950 to 0.988 | |
| Cyclosporine A (n = 44) | Intercept | -3.9830 | -14.07 to 4.396 |
| Slope | 0.9846 | 0.8929 to 1.129 | |
| Spearman’s ρ | 0.985 | 0.972 to 0.992 |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download