Dear Editor,
Shwachman-Diamond syndrome (SDS) is a rare multisystem disorder characterized by exocrine pancreatic dysfunction and hematologic abnormalities [1-3]. The phenotypic spectrum vary widely; therefore, genetic diagnosis is essential [1, 4, 5]. Approximately 90% of SDS cases are caused by SBDS pathogenic variants [4]. SBDS is located on chromosome 7, whereas pseudogene SBDSP1 with a highly homologous sequence (97% identical), located 5.8 Mb downstream, appears to be duplicated and inverted [1, 6, 7].
We report a patient with SDS. This study was approved by the Institutional Review Board of Seoul National University Hospital (SNUH), Korea (approval number: 2212-074-1385). Informed consent for genetic testing was obtained from the patient’s parents.
A 3-month-old female visited the SNUH on March 2022 with fever, bicytopenia (Hb=28 g/L, absolute neutrophil count=688 ×106/L), and failure to thrive. A bone marrow test showed normocellularity with a normal female karyotype. Elevated stool fatty acid, low serum amylase and lipase, continuously increasing AST and ALT levels were observed.
A multigene panel test was performed. This test revealed a homozygous SBDS variant (Fig. 1A): NM_016038.4(SBDS):c.258+2T>C p.? The total read count was 20 with a 100% variant allele frequency (VAF). We assessed the variant pathogenic based on the 2015 American College of Medical Genetics and Genomics and the Association for Molecular Pathology guidelines [8]. The trueness of this observation was doubted because the locus read depth was approximately half that obtained from other samples in the same capture pool, and the patient’s phenotype was not typical. We performed Sanger sequencing, using PCR primers specifically designed for SBDS. Sanger sequencing revealed another pathogenic variant (Fig. 1B), c.[183_184delinsCT;201A>G] p.(Lys62*), and a heterozygous state in the previously observed variant. The trans status of the two variants was verified by genomic DNA sequencing of both parents.
Through a literature review, we identified three reported cases with the same variant composition (Table 1) [6, 7]. We inferred that the VAF differed significantly among laboratories. Wu, et al. [7] reported a false-positive (FP) variant at the SBDSP1 paralogous sequence variant (PSV) locus, n.533+10T>C (NR_001588, represented as n.489T>C in Wu, et al. [7]). Wu, et al. [7] suggested that the next-generation sequencing (NGS) reads of SBDS may have been misaligned with those of SBDSP1, leading to observation of a variant at position c.141 (i.e., the FP variant n.424T>C). Peng, et al. [9] also reported a patient; however, this is not included in Table 1 because the read depth information were not provided. Nevertheless, a VAF of 70% for c.258+2C>T and false-negative (FN) calls of c.183_184delinsCT could be presumed based on the Integrative Genomics Viewer image provided. The reason for the different efficiencies of read mapping warrants further investigation as none of these studies examined this issue. Different alignment tools were used at the SNUH and in the previous studies, which might explain these differences.
Most population databases are based on short-read NGS results, which can lead to false allele frequencies for genes with pseudogenes. To establish the actual allele frequency of SBDS, we performed SBSD-specific sequencing using residual samples from 102 routine health check-up patients. We detected more altered alleles using Sanger sequencing than reported in the gnomAD or Korean reference database (Table 1). The allele frequencies we calculated for c.184A>T and c.201A>G, were significantly different from those in the gnomAD database for the Korean population (P=0.026 and P=0.001, respectively). c.258+2T>C was found to be 2.6-fold more prevalent in our study, but the difference was not significant (P=0.176).
In NGS reads, more variants at SBDS PSV loci increase the likelihood of misalignment at SBDSP1 loci, resulting in more FN results. Multiple mismatches within the c.[183_184delinsCT;201A>G] allele resulted in all reads being aligned to SBDSP1 loci.
Misalignments occur more frequently when a number of PSVs are exchanged in close proximity, as indicated by the conversion between the parent and its pseudogene [7]. Shortfalls have been acknowledged and efforts to overcome them are in progress. Alternatives include RNA-sequencing, long-read sequencing, and a tool to infer the haplotype [6, 9]. We recommend comparing the read depths of any suspicious variant detected within the patient and among the samples from the same capture pool in terms of the variant, and pseudogene loci where the reads might have been misaligned [7].
In summary, we report the genetic diagnosis of a patient with two common SBDS pathogenic variants. By comparing the VAFs of variants in previously reported cases, we demonstrate the challenges of short-read NGS in testing for pseudogene-related variants. Considering the composition of the control population allele frequency database, we established the Korean allele frequencies of four variants through SBDS-specific sequencing. The risk of erroneous diagnosis of disease-causing genes with pseudogenes when performing short-read NGS should be acknowledged.
Notes
REFERENCES
1. Nelson A, Myers K. Adam MP, Mirzaa GM, editors. 2008. Shwachman-Diamond Syndrome. GeneReviews®. University of Washington, Seattle;Seattle:
2. Furutani E, Liu S, Galvin A, Steltz S, Malsch MM, Loveless SK, et al. 2022; Hematologic complications with age in Shwachman-Diamond syndrome. Blood Adv. 6:297–306. DOI: 10.1182/bloodadvances.2021005539. PMID: 34758064. PMCID: PMC8753194.

3. Orphanet. The portal for rare diseases and orphan drugs. https://www.orpha.net/. Updated on June 2014.
4. Warren AJ. 2018; Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome. Adv Biol Regul. 67:109–27. DOI: 10.1016/j.jbior.2017.09.002. PMID: 28942353. PMCID: PMC6710477.

5. Lawal OS, Mathur N, Eapi S, Chowdhury R, Malik BH. 2020; Liver and cardiac involvement in Shwachman-Diamond syndrome: a literature review. Cureus. 12:e6676. DOI: 10.7759/cureus.6676.

6. Yamada M, Uehara T, Suzuki H, Takenouchi T, Inui A, Ikemiyagi M, et al. 2020; Shortfall of exome analysis for diagnosis of Shwachman-Diamond syndrome: mismapping due to the pseudogene SBDSP1. Am J Med Genet A. 182:1631–6. DOI: 10.1002/ajmg.a.61598. PMID: 32412173.
7. Wu D, Zhang L, Qiang Y, Wang K. 2022; Improved detection of SBDS gene mutation by a new method of next-generation sequencing analysis based on the Chinese mutation spectrum. PLoS One. 17:e0269029. DOI: 10.1371/journal.pone.0269029. PMID: 36512530. PMCID: PMC9747038.

8. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. 2015; Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–24. DOI: 10.1038/gim.2015.30. PMID: 25741868. PMCID: PMC4544753.

9. Peng X, Dong X, Wang Y, Wu B, Wang H, Lu W, et al. 2022; Overcoming the pitfalls of next-generation sequencing-based molecular diagnosis of Shwachman-Diamond syndrome. J Mol Diagn. 24:1240–53. DOI: 10.1016/j.jmoldx.2022.09.002. PMID: 36162759.

Fig. 1
Molecular findings in the present case. (A) Read alignments of NGS of the patient shown by the IGV software. (B) Sanger sequencing results of the patient and her parents. (C) Schema of SBDS and SBDSP1 conversion at long arm of chromosome 7. The sequence in black circles are derived from SBDS and those in white circles are from SBDSP1. Only 5 bases that are distinctive are described. The SBDS-specific primer set sequence and their approximate binding locus are indicated.
Abbreviations: NGS, next-generation sequencing; IGV, Integrative Genomics Viewer.
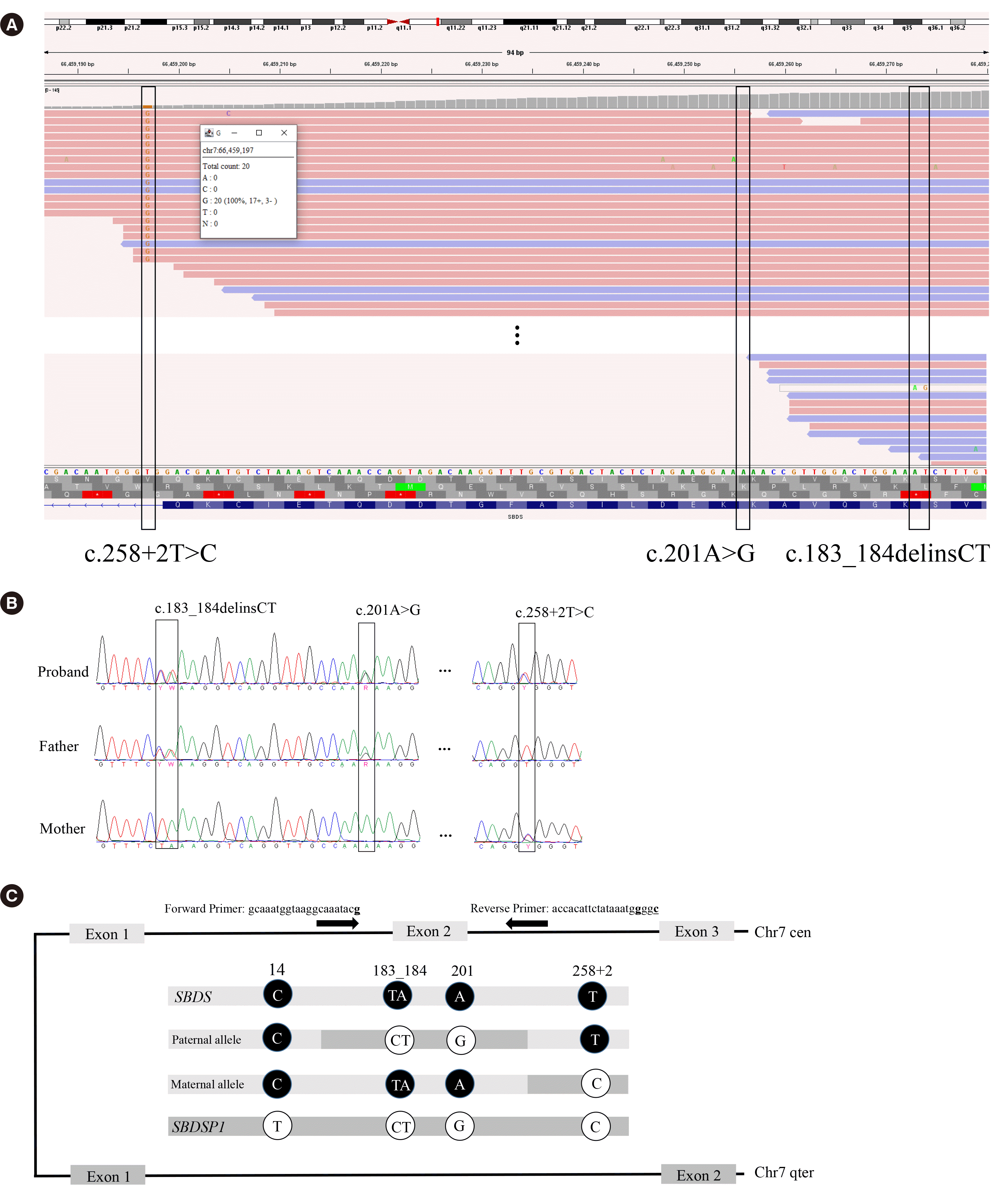
Table 1
Variant allele frequency and variant/total read counts of the three SBDS variants obtained from a multi-gene panel test of our patient and exome testing of two previously reported cases. Allele frequencies of SBDS variants within exon 2 and intron 2 inferred to have arisen from conversion between SBDS and SBDSP1
| Variant‡ | Genomic description§ | Variant allele frequency (variant read count/total read count) | Allele frequency (allele count/total allele number) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Our patient | Patient 1* | Patient 2* | Patient 3† | gnomAD (v.2.1.1.) | KRGDB¶ | This study** | ||||
|
|
||||||||||
| Total | East Asian | Korean | ||||||||
| c.141C >T, p.(Leu47 =) | Chr7:66459316 | N/A | 0.006781 (1,917/2,826,900) | 0.03356 (669/19,936) | 0.03230 (123/3,808) | 0.028247 | 0.034314 (7/204) | |||
| c.184A >T, p.(Lys62*)†† | Chr7:66459273 | 0% (1/106) | 5% (3/56) | 3% (2/68) | 9% (3/35) | 0.0002582 (73/282,686) | 0.0004514 (9/19,936) | 0.0005241 (2/3,816) | 0.002058 | 0.004902 (1/204) |
| c.201A > G, p.(Lys67 =) | Chr7:66459256 | 0% (0/93) | 7% (4/56) | 4% (3/71) | 10% (3/31) | 0.08803 (24,881/282,658) | 0.003262 (65/19,928) | 0.003673 (14/3,812) | 0.004077 | 0.019608 (4/204) |
| c.258+2T > C, p.? | Chr7:66459197 | 100% (20/20) | 87% (39/45) | 85% (50/59) | 43% (15/35) | 0.003879 (1,096/282,568) | 0.005175 (103/19,902) | 0.003675 (14/3,810) | 0.005824 | 0.009804 (2/204) |
*From Yamada, et al. [6]; †From Wu, et al. [7]; ‡NM_016038.4; §Based on the GRCh37/hg19 reference; ¶All-merged group (1,722 patients) from all phases; **Based on Sanger sequencing results of 102 Koreans; ††Described as c.184A>T, p.(Lys62*) in the table in contrast to c.183_184delinsCT in the text because c.183T>C, p.(Ser61=) is predicted not to cause any protein change.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download