Abstract
Notes
AUTHOR CONTRIBUTIONS
Shin JH designed the study; Choi MJ and Byun SA performed the laboratory measurements and molecular studies; Kim MN, Lee WG, Lee J, Yong D, Chang CL, Won EJ, and Kim SH collected the clinical isolates and data; Shin JH, Kwon YJ, and Lee SY wrote the preliminary manuscript; Shin JH, Kwon YJ, and Lee SY analyzed the data; Shin JH revised the manuscript; Kim MN, Lee WG, Lee J, Chang CL, Won EJ, and Kim SH provided valuable comments and recommendations. All authors revised and accepted the final version of the manuscript.
REFERENCES
Fig. 1
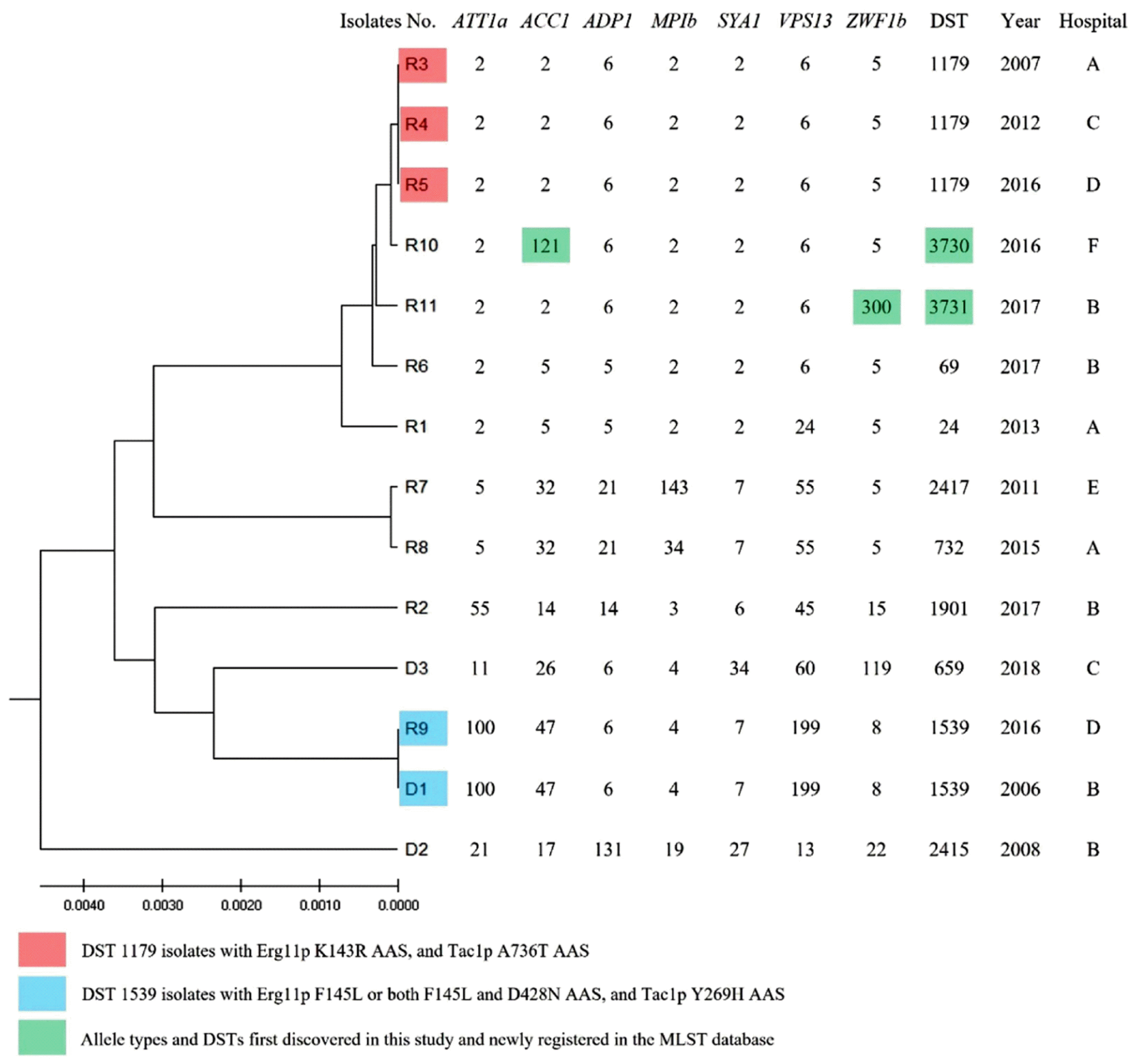
Table 1
| Isolate No. | MIC (μg/mL)* | Erg11p AAS found in† | Tac1p AAS found in† | Mrr1p AAS found in† | Upc2p AAS found in† | ||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||||
| FLC/VOR/ITRA/POSA | FNS isolates only‡ | Both FNS and FS isolates | FNS isolates only‡ | Both FNS and FS isolates|| | FNS isolates only | Both FNS and FS isolates|| | FNS isolates only | Both FNS and FS isolates | |
| R1 | > 256/> 8/> 16/>8 | None | D116E, K128T | L744I§ | N772K, S935L | None | L171P, L248V, V341E | None | None |
| R2 | > 256/> 8/> 16/>8 | K143R | D116E, K128T | None | I558V, N772K | None | - | None | I142S |
| R3 | 16/0.12/0.25/0.25 | K143R | D116E, K128T | A736T | N772K, S935L | None | L171P, L248V, V341E | None | None |
| R4 | 16/0.12/0.25/0.25 | K143R | D116E, K128T | A736T | N772K, S935L | None | L171P, L248V, V341E | None | None |
| R5 | 16/0.12/0.25/0.25 | K143R | D116E, K128T | A736T | N772K, S935L | None | N159H, I160T, A162P, L171P, L248V, V341E | None | None |
| R6 | 16/0.5/0.5/0.5 | R264T§, G464S | D116E, K128T | N972K§ | N772K, S935L | None | V27I¶, L171P, L248V, V341E | None | None |
| R7 | 8/0.12/0.25/0.25 | None | D116E, E266D, V488I | R673L | I895T, N896S | None | L171P, V341E | None | None |
| R8 | 8/0.12/0.06/0.03 | None | D116E, E266D, V488I | None | I895T, N896S | None | L171P, V341E | None | None |
| R9 | 8/0.12/0.25/0.25 | F145L | K342R¶ | Y269H§ | N896S | None | L171P, V341E | None | I142S |
| R10 | 8/0.12/0.25/0.25 | None | D116E, K128T | A736V | N772K, S935L | None | N159H, I160T, A162P, L171P, L248V, V341E | None | None |
| R11 | 8/0.03/0.12/0.06 | K143R | D116E, K128T | A736T | N772K, S935L | None | N159H, I160T, A162P, L171P, L248V, V341E | None | None |
| D1 | 4/0.12/0.25/0.25 | F145L, D428N§ | K342R¶ | Y269H§ | N896S | N33S§ | L171P, V341E | None | I142S, P299L |
| D2 | 4/0.015/0.06/0.03 | None | E266D, V437I | None | K87N, M170I, N174D, F189S | None | L171P, L248K, V341E | None | I142S |
| D3 | 4/0.06/0.25/0.25 | None | E266D, V488I | T225A | K87N, M170I, N174D, F189S, N772K, N896S | None | - | None | I142S, S190N, S228N |
| S1 | 0.5/0.03/0.03/0.015 | None | D153E | None | I895T, N896S | None | L171P | None | R68K, I142S, S228N |
| S2 | 0.5/0.03/0.06/0.03 | None | E266D, V488I | None | N896S | None | S16I, T73K, L171P | None | None |
| S3 | 0.25/0.008/0.015/0.015 | None | D116E, D153E | None | I895T, N896S | None | L171P | None | R68K, I142S, S228N |
| S4 | 0.25/0.015/0.03/0.015 | None | D116E, D153E | None | I895T, N896S | None | L171P | None | I142S, S228N |
| S5 | 0.25/0.008/0.03/0.03 | None | E266D, V437I | None | K87N, A90T, M170I, N174D, F189S, F222L | None | P19L, G75R, N937K, F1032L | None | R68K, I142S, S190N, S228N |
| S6 | 0.25/0.008/0.03/0.015 | None | E266D, V437I | None | M170I, N174D, F189S | None | N937K, F1032L | None | R68K, I142S, S190N, S228N |
| S7 | 0.25/0.008/0.03/0.015 | None | E266D, V437I | None | A90T | None | N937K, F1032L | None | R68K, I142S, S190N, S228N |
| S8 | 0.25/0.015/0.03/0.03 | None | D116E, D153E | None | I895T, N896S | None | - | None | R68K, I142S, S288N, P299L, A300P |
| S9 | 0.25/0.015/0.03/0.03 | None | D116E, K128T | None | N772K | None | L171P, L248V, V341E | None | None |
| S10 | 0.25/0.008/0.015/0.015 | None | E266D, V488I | None | N772K, N896S | None | L171P | None | None |
| S11 | 0.25/0.008/0.015/0.03 | None | E266D, V488I | None | N896S | None | L171P, S219F | None | None |
| S12 | 0.25/0.008/0.03/0.015 | None | D116E | None | I895T, N896S | None | L171P | None | I142S, S228N |
*Antifungal MICs were determined using the Sensititre Yeast One system (Thermo Fisher Scientific Inc., Cleveland, OH, USA); †The sequences of the isolates were compared and analyzed based on the reference sequences for ERG11 (GenBank accession No. X13296), TAC1 (GenBank accession No. DQ393587), MRR1 (GenBank accession no. XM711520), and UPC2 (GenBank accession No. EU583451) from C. albicans [6]; homozygous alleles are underlined; ‡AASs that were previously detected in fluconazole-resistant C. albicans isolates are shown in bold; §New AASs (Erg11p R264T and D428N AASs, Tac1p Y269H, N744I, and N972K, and Mrr1p N33S) that were found in the FNS isolates of C. albicans in this study have been deposited into GenBank with accession numbers OQ161592, OQ161593, OQ161595, OQ383350, OQ161594, and OQ161598, respectively; ||Eight common Tac1p AASs (F104V, S199N, R206H, V207A, N396S, D776N, E829Q, and L941P) and one Mrr1p ASS (E1020Q) that were found in almost all (≥23) isolates were excluded; ¶Erg11p K342R AAS and Mrr1p V27I AAS were reported previously in FS isolates [7, 17].
Table 2
| Isolate No. | Age (yr)/sex | Diagnosis | Prior antifungal exposure | Immuno-suppression | CVC | Duration of fungemia (days) | Antifungal treatment | Patient outcome (days)* |
|---|---|---|---|---|---|---|---|---|
| R1 | 30/F | Acute myeloid leukemia | Yes (AMB) | Yes | Yes | 5 | AMB, ANI | Death (6) |
| R2 | 63/F | Pancreatic cancer | No | Yes | Yes | 1 | CAS | Death (52) |
| R3 | 52/M | Diabetes mellitus | No | No | No | 10 | AMB | Improved |
| R4 | 75/M | COPD | No | No | No | 3 | FLC | Death (19) |
| R5 | 49/F | Breast cancer | No | No | Yes | 1 | None | Death (2) |
| R6 | 52/M | T/NK-cell lymphoma | No | No | Yes | 5 | CAS | Death (7) |
| R7 | 74/M | Diabetes mellitus | No | No | Yes | 1 | None | Death (1) |
| R8 | 74/F | Chronic myeloid leukemia | Yes (FLC) | Yes | No | 2 | CAS | Death (4) |
| R9 | 62/M | Rheumatoid arthritis | No | No | Yes | 4 | MICA | Improved |
| R10 | 79/F | Traumatic subdural hemorrhage | No | No | Yes | 1 | FLC | Improved |
| R11 | 80/M | Spinal abscess | No | No | Yes | 2 | CAS | Improved |
| D1 | 79/M | Diabetes mellitus | Yes (AMB) | No | Yes | 8 | FLC | Death (15) |
| D2 | 43/M | Down syndrome | No | Yes | Yes | 6 | AMB | Improved |
| D3 | 67/F | Fulminant myocarditis, COPD | No | No | Yes | 1 | None | Death (3) |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download