1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021; 71:209–49.
2. Wang X, Zhang H, Chen X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019; 2:141–60.
3. Ruth JR, Pant DK, Pan TC, Seidel HE, Baksh SC, Keister BA, et al. Cellular dormancy in minimal residual disease following targeted therapy. Breast Cancer Res. 2021; 23:63.
4. Tachtsidis A, McInnes LM, Jacobsen N, Thompson EW, Saunders CM. Minimal residual disease in breast cancer: an overview of circulating and disseminated tumour cells. Clin Exp Metastasis. 2016; 33:521–50.

5. Kang Y, Pantel K. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer Cell. 2013; 23:573–81.
6. Badia-Ramentol J, Linares J, Gomez-Llonin A, Calon A. Minimal residual disease, metastasis and immunity. Biomolecules. 2021; 11:130.

7. Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells: mechanisms of immune surveillance and escape. Nat Rev Clin Oncol. 2017; 14:155–67.
8. Yeh AC, Ramaswamy S. Mechanisms of cancer cell dormancy: another hallmark of cancer? Cancer Res. 2015; 75:5014–22.
9. Endo H, Inoue M. Dormancy in cancer. Cancer Sci. 2019; 110:474–80.
10. Goss PE, Chambers AF. Does tumour dormancy offer a therapeutic target? Nat Rev Cancer. 2010; 10:871–7.

11. Damen MP, van Rheenen J, Scheele C. Targeting dormant tumor cells to prevent cancer recurrence. FEBS J. 2021; 288:6286–303.

12. Recasens A, Munoz L. Targeting cancer cell dormancy. Trends Pharmacol Sci. 2019; 40:128–41.
13. Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014; 14:611–22.

14. Phan TG, Croucher PI. The dormant cancer cell life cycle. Nat Rev Cancer. 2020; 20:398–411.

15. Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007; 7:834–46.

16. Indraccolo S, Stievano L, Minuzzo S, Tosello V, Esposito G, Piovan E, et al. Interruption of tumor dormancy by a transient angiogenic burst within the tumor microenvironment. Proc Natl Acad Sci U S A. 2006; 103:4216–21.
17. Swann JB, Smyth MJ. Immune surveillance of tumors. J Clin Invest. 2007; 117:1137–46.

18. Aqbi HF, Wallace M, Sappal S, Payne KK, Manjili MH. IFN-gamma orchestrates tumor elimination, tumor dormancy, tumor escape, and progression. J Leukoc Biol. 2018; 103:1219–23.
19. Correia AL, Guimaraes JC, Auf der Maur P, De Silva D, Trefny MP, Okamoto R, et al. Hepatic stellate cells suppress NK cell-sustained breast cancer dormancy. Nature. 2021; 594:566–71.

20. Liu Y, Liang X, Yin X, Lv J, Tang K, Ma J, et al. Blockade of IDO-kynurenine-AhR metabolic circuitry abrogates IFN-gamma-induced immunologic dormancy of tumor-repopulating cells. Nat Commun. 2017; 8:15207.
21. Muller-Hermelink N, Braumuller H, Pichler B, Wieder T, Mailhammer R, Schaak K, et al. TNFR1 signaling and IFN-gamma signaling determine whether T cells induce tumor dormancy or promote multistage carcinogenesis. Cancer Cell. 2008; 13:507–18.
22. Braumuller H, Wieder T, Brenner E, Assmann S, Hahn M, Alkhaled M, et al. T-helper-1-cell cytokines drive cancer into senescence. Nature. 2013; 494:361–5.
23. Basu S, Dong Y, Kumar R, Jeter C, Tang DG. Slow-cycling (dormant) cancer cells in therapy resistance, cancer relapse and metastasis. Semin Cancer Biol. 2022; 78:90–103.
24. Santos-de-Frutos K, Djouder N. When dormancy fuels tumour relapse. Commun Biol. 2021; 4:747.

25. Townson JL, Chambers AF. Dormancy of solitary metastatic cells. Cell Cycle. 2006; 5:1744–50.

26. Spiliotaki M, Mavroudis D, Kapranou K, Markomanolaki H, Kallergi G, Koinis F, et al. Evaluation of proliferation and apoptosis markers in circulating tumor cells of women with early breast cancer who are candidates for tumor dormancy. Breast Cancer Res. 2014; 16:485.

27. Gilje B, Nordgard O, Tjensvoll K, Janssen EA, Soiland H, Smaaland R, et al. Mitotic activity and bone marrow micrometastases have independent prognostic value in node positive breast cancer patients. Breast Cancer Res Treat. 2011; 128:137–46.

28. Kato TA, Haskins JS. Mitotic index analysis. Methods Mol Biol. 2023; 2519:17–26.

29. Oki T, Nishimura K, Kitaura J, Togami K, Maehara A, Izawa K, et al. A novel cell-cycle-indicator, mVenus-p27K-, identifies quiescent cells and visualizes G0–G1 transition. Sci Rep. 2014; 4:4012.

30. Moore N, Houghton J, Lyle S. Slow-cycling therapy-resistant cancer cells. Stem Cells Dev. 2012; 21:1822–30.

31. Yumoto K, Berry JE, Taichman RS, Shiozawa Y. A novel method for monitoring tumor proliferation in vivo using fluorescent dye DiD. Cytometry A. 2014; 85:548–55.

32. Chauvistre H, Shannan B, Daignault-Mill SM, Ju RJ, Picard D, Egetemaier S, et al. Persister state-directed transitioning and vulnerability in melanoma. Nat Commun. 2022; 13:3055.
33. Tario JD Jr, Humphrey K, Bantly AD, Muirhead KA, Moore JS, Wallace PK. Optimized staining and proliferation modeling methods for cell division monitoring using cell tracking dyes. J Vis Exp. 2012; e4287.
34. Ebinger S, Ozdemir EZ, Ziegenhain C, Tiedt S, Castro Alves C, Grunert M, et al. Characterization of rare, dormant, and therapy-resistant cells in acute lymphoblastic leukemia. Cancer Cell. 2016; 30:849–62.
35. Preciado JA, Aksan A. Method to isolate dormant cancer cells from heterogeneous populations. Methods Mol Biol. 2022; 2394:19–29.

36. Pradhan S, Sperduto JL, Farino CJ, Slater JH. Engineered in vitro models of tumor dormancy and reactivation. J Biol Eng. 2018; 12:37.

37. Gu Y, Bui T, Muller WJ. Exploiting mouse models to recapitulate clinical tumor dormancy and recurrence in breast cancer. Endocrinology. 2022; 163:bqac055.
38. Sosa MS, Avivar-Valderas A, Bragado P, Wen HC, Aguirre-Ghiso JA. ERK1/2 and p38alpha/beta signaling in tumor cell quiescence: opportunities to control dormant residual disease. Clin Cancer Res. 2011; 17:5850–7.
39. Aguirre-Ghiso JA, Estrada Y, Liu D, Ossowski L. ERK(MAPK) activity as a determinant of tumor growth and dormancy; regulation by p38(SAPK). Cancer Res. 2003; 63:1684–95.

40. Barney LE, Hall CL, Schwartz AD, Parks AN, Sparages C, Galarza S, et al. Tumor cell-organized fibronectin maintenance of a dormant breast cancer population. Sci Adv. 2020; 6:eaaz4157.

41. Rehman SK, Haynes J, Collignon E, Brown KR, Wang Y, Nixon AM, et al. Colorectal cancer cells enter a diapause-like DTP state to survive chemotherapy. Cell. 2021; 184:226–42.
42. Cho J, Min HY, Lee HJ, Hyun SY, Sim JY, Noh M, et al. RGS2-mediated translational control mediates cancer cell dormancy and tumor relapse. J Clin Invest. 2021; 131:e136779.

43. Pranzini E, Raugei G, Taddei ML. Metabolic features of tumor dormancy: possible therapeutic strategies. Cancers (Basel). 2022; 14:547.

44. Valcourt JR, Lemons JM, Haley EM, Kojima M, Demuren OO, Coller HA. Staying alive: metabolic adaptations to quiescence. Cell Cycle. 2012; 11:1680–96.
45. Ranganathan AC, Adam AP, Zhang L, Aguirre-Ghiso JA. Tumor cell dormancy induced by p38SAPK and ER-stress signaling: an adaptive advantage for metastatic cells? Cancer Biol Ther. 2006; 5:729–35.

46. Evertts AG, Manning AL, Wang X, Dyson NJ, Garcia BA, Coller HA. H4K20 methylation regulates quiescence and chromatin compaction. Mol Biol Cell. 2013; 24:3025–37.

47. Milanovic M, Fan DN, Belenki D, Dabritz JH, Zhao Z, Yu Y, et al. Senescence-associated reprogramming promotes cancer stemness. Nature. 2018; 553:96–100.

48. Truskowski K, Amend SR, Pienta KJ. Dormant cancer cells: programmed quiescence, senescence, or both? Cancer Meta-stasis Rev. 2023; 42:37–47.
49. Schmitt CA, Wang B, Demaria M. Senescence and cancer: role and therapeutic opportunities. Nat Rev Clin Oncol. 2022; 19:619–36.
50. Iwasa H, Han J, Ishikawa F. Mitogen-activated protein kinase p38 defines the common senescence-signalling pathway. Genes Cells. 2003; 8:131–44.

51. Prunier C, Alay A, van Dijk M, Ammerlaan KL, van Gelderen S, Marvin DL, et al. Breast cancer dormancy is associated with a 4NG1 state and not senescence. NPJ Breast Cancer. 2021; 7:140.
52. Triana-Martinez F, Loza MI, Dominguez E. Beyond tumor suppression: senescence in cancer stemness and tumor dormancy. Cells. 2020; 9:346.

53. Saleh T, Tyutyunyk-Massey L, Gewirtz DA. Tumor cell escape from therapy-induced senescence as a model of disease recurrence after dormancy. Cancer Res. 2019; 79:1044–6.

54. Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015; 16:225–38.

55. Desai A, Yan Y, Gerson SL. Concise reviews: cancer stem cell targeted therapies: toward clinical success. Stem Cells Transl Med. 2019; 8:75–81.

56. Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells: what challenges do they pose? Nat Rev Drug Discov. 2014; 13:497–512.

57. Medema JP. Cancer stem cells: the challenges ahead. Nat Cell Biol. 2013; 15:338–44.
58. Lambert AW, Weinberg RA. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat Rev Cancer. 2021; 21:325–38.

59. Weidenfeld K, Barkan D. EMT and stemness in tumor dormancy and outgrowth: are they intertwined processes? Front Oncol. 2018; 8:381.
60. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008; 133:704–15.
61. Pradella D, Naro C, Sette C, Ghigna C. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer. 2017; 16:8.

62. Wang SS, Jiang J, Liang XH, Tang YL. Links between cancer stem cells and epithelial-mesenchymal transition. Onco Targets Ther. 2015; 8:2973–80.
63. Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012; 22:709–24.

64. Jia Q, Yang F, Huang W, Zhang Y, Bao B, Li K, et al. Low Levels of Sox2 are required for melanoma tumor-repopulating cell dormancy. Theranostics. 2019; 9:424–35.

65. Ohta Y, Fujii M, Takahashi S, Takano A, Nanki K, Matano M, et al. Cell-matrix interface regulates dormancy in human colon cancer stem cells. Nature. 2022; 608:784–94.

66. Zhou N, Wu X, Yang B, Yang X, Zhang D, Qing G. Stem cell characteristics of dormant cells and cisplatin-induced effects on the stemness of epithelial ovarian cancer cells. Mol Med Rep. 2014; 10:2495–504.

67. Fischer M, Schade AE, Branigan TB, Muller GA, DeCaprio JA. Coordinating gene expression during the cell cycle. Trends Biochem Sci. 2022; 47:1009–22.

68. Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006; 25:5220–7.

69. Kwon JS, Everetts NJ, Wang X, Wang W, Della Croce K, Xing J, et al. Controlling depth of cellular quiescence by an Rb-E2F network switch. Cell Rep. 2017; 20:3223–35.

70. Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013; 13:585–95.

71. Litovchick L, Florens LA, Swanson SK, Washburn MP, DeCaprio JA. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 2011; 25:801–13.

72. MacDonald J, Ramos-Valdes Y, Perampalam P, Litovchick L, DiMattia GE, Dick FA. A systematic analysis of negative growth control implicates the DREAM complex in cancer cell dormancy. Mol Cancer Res. 2017; 15:371–81.

73. Guiley KZ, Liban TJ, Felthousen JG, Ramanan P, Litovchick L, Rubin SM. Structural mechanisms of DREAM complex assembly and regulation. Genes Dev. 2015; 29:961–74.

74. Schade AE, Oser MG, Nicholson HE, DeCaprio JA. Cyclin D-CDK4 relieves cooperative repression of proliferation and cell cycle gene expression by DREAM and RB. Oncogene. 2019; 38:4962–76.
75. Boichuk S, Parry JA, Makielski KR, Litovchick L, Baron JL, Zewe JP, et al. The DREAM complex mediates GIST cell quiescence and is a novel therapeutic target to enhance imatinib-induced apoptosis. Cancer Res. 2013; 73:5120–9.

76. Wang P, Karakose E, Argmann C, Wang H, Balev M, Brody RI, et al. Disrupting the DREAM complex enables proliferation of adult human pancreatic beta cells. J Clin Invest. 2022; 132:e157086.
77. Kim MJ, Cervantes C, Jung YS, Zhang X, Zhang J, Lee SH, et al. PAF remodels the DREAM complex to bypass cell quiescence and promote lung tumorigenesis. Mol Cell. 2021; 81:1698–714.

78. Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010; 123:4195–200.

79. Yue B. Biology of the extracellular matrix: an overview. J Glaucoma. 2014; 23(8 Suppl 1):S20–3.
80. Hynes RO, Naba A. Overview of the matrisome: an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol. 2012; 4:a004903.
81. Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 2016; 49:10–24.

82. Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun. 2020; 11:5120.

83. Wei Y, Lukashev M, Simon DI, Bodary SC, Rosenberg S, Doyle MV, et al. Regulation of integrin function by the urokinase receptor. Science. 1996; 273:1551–5.

84. Aguirre Ghiso JA, Kovalski K, Ossowski L. Tumor dormancy induced by downregulation of urokinase receptor in human carcinoma involves integrin and MAPK signaling. J Cell Biol. 1999; 147:89–104.

85. Aguirre-Ghiso JA, Liu D, Mignatti A, Kovalski K, Ossow-ski L. Urokinase receptor and fibronectin regulate the ERK(MAPK) to p38(MAPK) activity ratios that determine carcinoma cell proliferation or dormancy in vivo. Mol Biol Cell. 2001; 12:863–79.
86. Barkan D, Kleinman H, Simmons JL, Asmussen H, Kamaraju AK, Hoenorhoff MJ, et al. Inhibition of metastatic outgrowth from single dormant tumor cells by targeting the cytoskeleton. Cancer Res. 2008; 68:6241–50.

87. Barkan D, El Touny LH, Michalowski AM, Smith JA, Chu I, Davis AS, et al. Metastatic growth from dormant cells induced by a col-I-enriched fibrotic environment. Cancer Res. 2010; 70:5706–16.

88. Cho J, Lee HJ, Hwang SJ, Min HY, Kang HN, Park AY, et al. The interplay between slow-cycling, chemoresistant cancer cells and fibroblasts creates a proinflammatory niche for tumor progression. Cancer Res. 2020; 80:2257–72.

89. Keeratichamroen S, Lirdprapamongkol K, Svasti J. Mechanism of ECM-induced dormancy and chemoresistance in A549 human lung carcinoma cells. Oncol Rep. 2018; 39:1765–74.

90. Di Martino JS, Nobre AR, Mondal C, Taha I, Farias EF, Fertig EJ, et al. A tumor-derived type III collagen-rich ECM niche regulates tumor cell dormancy. Nat Cancer. 2022; 3:90–107.

91. Almog N, Ma L, Raychowdhury R, Schwager C, Erber R, Short S, et al. Transcriptional switch of dormant tumors to fast-growing angiogenic phenotype. Cancer Res. 2009; 69:836–44.

92. Kazerounian S, Yee KO, Lawler J. Thrombospondins in cancer. Cell Mol Life Sci. 2008; 65:700–12.
93. Rouanne M, Adam J, Goubar A, Robin A, Ohana C, Louvet E, et al. Osteopontin and thrombospondin-1 play opposite roles in promoting tumor aggressiveness of primary resected non-small cell lung cancer. BMC Cancer. 2016; 16:483.

94. Boyerinas B, Zafrir M, Yesilkanal AE, Price TT, Hyjek EM, Sipkins DA. Adhesion to osteopontin in the bone marrow niche regulates lymphoblastic leukemia cell dormancy. Blood. 2013; 121:4821–31.

95. Parker AL, Cox TR. The role of the ECM in lung cancer dormancy and outgrowth. Front Oncol. 2020; 10:1766.

96. Rodrigues LR, Teixeira JA, Schmitt FL, Paulsson M, Lindmark-Mansson H. The role of osteopontin in tumor progression and metastasis in breast cancer. Cancer Epidemiol Biomarkers Prev. 2007; 16:1087–97.
97. Jiang Y, Zhang H, Wang J, Liu Y, Luo T, Hua H. Targeting extracellular matrix stiffness and mechanotransducers to improve cancer therapy. J Hematol Oncol. 2022; 15:34.

98. Doue M, Okwieka A, Berquand A, Gorisse L, Maurice P, Velard F, et al. Carbamylation of elastic fibers is a molecular substratum of aortic stiffness. Sci Rep. 2021; 11:17827.
99. Schrader J, Gordon-Walker TT, Aucott RL, van Deemter M, Quaas A, Walsh S, et al. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology. 2011; 53:1192–205.

100. Kondapaneni RV, Rao SS. Matrix stiffness and cluster size collectively regulate dormancy versus proliferation in brain metastatic breast cancer cell clusters. Biomater Sci. 2020; 8:6637–46.
101. Anlas AA, Nelson CM. Soft microenvironments induce chemoresistance by increasing autophagy downstream of integrin-linked kinase. Cancer Res. 2020; 80:4103–13.
102. Liu Y, Lv J, Liang X, Yin X, Zhang L, Chen D, et al. Fibrin stiffness mediates dormancy of tumor-repopulating cells via a Cdc42-driven Tet2 epigenetic program. Cancer Res. 2018; 78:3926–37.

103. Sistigu A, Musella M, Galassi C, Vitale I, De Maria R. Tuning cancer fate: tumor microenvironment’s role in cancer stem cell quiescence and reawakening. Front Immunol. 2020; 11:2166.

104. Butturini E, Carcereri de Prati A, Boriero D, Mariotto S. Tumor dormancy and interplay with hypoxic tumor microenvironment. Int J Mol Sci. 2019; 20:4305.

105. Ju S, Wang F, Wang Y, Ju S. CSN8 is a key regulator in hypoxia-induced epithelial-mesenchymal transition and dormancy of colorectal cancer cells. Mol Cancer. 2020; 19:168.
106. Ameri K, Jahangiri A, Rajah AM, Tormos KV, Nagarajan R, Pekmezci M, et al. HIGD1A regulates oxygen consumption, ROS production, and AMPK activity during glucose deprivation to modulate cell survival and tumor growth. Cell Rep. 2015; 10:891–9.

107. Bildik G, Liang X, Sutton MN, Bast RC Jr, Lu Z. DIRAS3: an imprinted tumor suppressor gene that regulates RAS and PI3K-driven cancer growth, motility, autophagy, and tumor dormancy. Mol Cancer Ther. 2022; 21:25–37.

108. Sutton MN, Huang GY, Zhou J, Mao W, Langley R, Lu Z, et al. Amino acid deprivation-induced autophagy requires upregulation of DIRAS3 through reduction of E2F1 and E2F4 transcriptional repression. Cancers (Basel). 2019; 11:603.

109. Wang L, Shang Z, Zhou Y, Hu X, Chen Y, Fan Y, et al. Autophagy mediates glucose starvation-induced glioblastoma cell quiescence and chemoresistance through coordinating cell metabolism, cell cycle, and survival. Cell Death Dis. 2018; 9:213.

110. Zhang M, Peng R, Wang H, Yang Z, Zhang H, Zhang Y, et al. Nanog mediated by FAO/ACLY signaling induces cellular dormancy in colorectal cancer cells. Cell Death Dis. 2022; 13:159.

111. Dong X, Xue H, Mo F, Lin YY, Lin D, Wong NKY, et al. Modeling androgen deprivation therapy-induced prostate cancer dormancy and its clinical implications. Mol Cancer Res. 2022; 20:782–93.

112. Kurppa KJ, Liu Y, To C, Zhang T, Fan M, Vajdi A, et al. Treatment-induced tumor dormancy through YAP-mediated transcriptional reprogramming of the apoptotic pathway. Cancer Cell. 2020; 37:104–22.

113. Min M, Spencer SL. Spontaneously slow-cycling subpopulations of human cells originate from activation of stress-response pathways. PLoS Biol. 2019; 17:e3000178.

114. Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009; 9:537–49.

115. Adam AP, George A, Schewe D, Bragado P, Iglesias BV, Ranganathan AC, et al. Computational identification of a p38SAPK-regulated transcription factor network required for tumor cell quiescence. Cancer Res. 2009; 69:5664–72.

116. Sosa MS, Parikh F, Maia AG, Estrada Y, Bosch A, Bragado P, et al. NR2F1 controls tumour cell dormancy via SOX9- and RARbeta-driven quiescence programmes. Nat Commun. 2015; 6:6170.
117. Cai B, Chang SH, Becker EB, Bonni A, Xia Z. p38 MAP kinase mediates apoptosis through phosphorylation of BimEL at Ser-65. J Biol Chem. 2006; 281:25215–22.

118. Zhuang S, Demirs JT, Kochevar IE. p38 mitogen-activated protein kinase mediates bid cleavage, mitochondrial dysfunction, and caspase-3 activation during apoptosis induced by singlet oxygen but not by hydrogen peroxide. J Biol Chem. 2000; 275:25939–48.

119. Sui X, Kong N, Ye L, Han W, Zhou J, Zhang Q, et al. p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett. 2014; 344:174–9.
120. Kroeger H, Grimsey N, Paxman R, Chiang WC, Plate L, Jones Y, et al. The unfolded protein response regulator ATF6 promotes mesodermal differentiation. Sci Signal. 2018; 11:eaan5785.

121. Fu Y, Li J, Lee AS. GRP78/BiP inhibits endoplasmic reticulum BIK and protects human breast cancer cells against estrogen starvation-induced apoptosis. Cancer Res. 2007; 67:3734–40.

122. Schewe DM, Aguirre-Ghiso JA. ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proc Natl Acad Sci U S A. 2008; 105:10519–24.
123. Gawrzak S, Rinaldi L, Gregorio S, Arenas EJ, Salvador F, Urosevic J, et al. MSK1 regulates luminal cell differentiation and metastatic dormancy in ER(+) breast cancer. Nat Cell Biol. 2018; 20:211–21.
124. Aman Y, Schmauck-Medina T, Hansen M, Morimoto RI, Simon AK, Bjedov I, et al. Autophagy in healthy aging and disease. Nat Aging. 2021; 1:634–50.

125. Akkoc Y, Peker N, Akcay A, Gozuacik D. Autophagy and cancer dormancy. Front Oncol. 2021; 11:627023.

126. Vera-Ramirez L. Cell-intrinsic survival signals. The role of autophagy in metastatic dissemination and tumor cell dormancy. Semin Cancer Biol. 2020; 60:28–40.

127. Chen Y, Gibson SB. Three dimensions of autophagy in regulating tumor growth: cell survival/death, cell proliferation, and tumor dormancy. Biochim Biophys Acta Mol Basis Dis. 2021; 1867:166265.

128. Gupta A, Roy S, Lazar AJ, Wang WL, McAuliffe JC, Reynoso D, et al. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (GIST). Proc Natl Acad Sci U S A. 2010; 107:14333–8.

129. Vera-Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun. 2018; 9:1944.

130. Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare S, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008; 118:3917–29.
131. Lu Z, Baquero MT, Yang H, Yang M, Reger AS, Kim C, et al. DIRAS3 regulates the autophagosome initiation complex in dormant ovarian cancer cells. Autophagy. 2014; 10:1071–92.

132. Weiss A, Attisano L. The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol. 2013; 2:47–63.

133. Bragado P, Estrada Y, Parikh F, Krause S, Capobianco C, Farina HG, et al. TGF-beta2 dictates disseminated tumour cell fate in target organs through TGF-beta-RIII and p38alpha/beta signalling. Nat Cell Biol. 2013; 15:1351–61.
134. Yumoto K, Eber MR, Wang J, Cackowski FC, Decker AM, Lee E, et al. Axl is required for TGF-beta2-induced dormancy of prostate cancer cells in the bone marrow. Sci Rep. 2016; 6:36520.
135. Kobayashi A, Okuda H, Xing F, Pandey PR, Watabe M, Hirota S, et al. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J Exp Med. 2011; 208:2641–55.
136. Pollak MN, Schernhammer ES, Hankinson SE. Insulin-like growth factors and neoplasia. Nat Rev Cancer. 2004; 4:505–18.

137. Rajbhandari N, Lin WC, Wehde BL, Triplett AA, Wagner KU. Autocrine IGF1 signaling mediates pancreatic tumor cell dormancy in the absence of oncogenic drivers. Cell Rep. 2017; 18:2243–55.
138. Shimizu T, Sugihara E, Yamaguchi-Iwai S, Tamaki S, Koyama Y, Kamel W, et al. IGF2 preserves osteosarcoma cell survival by creating an autophagic state of dormancy that protects cells against chemotherapeutic stress. Cancer Res. 2014; 74:6531–41.

139. Worster DT, Schmelzle T, Solimini NL, Lightcap ES, Millard B, Mills GB, et al. Akt and ERK control the proliferative response of mammary epithelial cells to the growth factors IGF-1 and EGF through the cell cycle inhibitor p57Kip2. Sci Signal. 2012; 5:ra19.
140. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013; 20:300–7.

141. Ruksha TG. MicroRNAs’ control of cancer cell dormancy. Cell Div. 2019; 14:11.

142. Nishikawa S, Dewi DL, Ishii H, Konno M, Haraguchi N, Kano Y, et al. Transcriptomic study of dormant gastrointestinal cancer stem cells. Int J Oncol. 2012; 41:979–84.
143. Almog N, Briggs C, Beheshti A, Ma L, Wilkie KP, Rietman E, et al. Transcriptional changes induced by the tumor dormancy-associated microRNA-190. Transcription. 2013; 4:177–91.

144. Vera-Ramirez L, Hunter KW. Tumor cell dormancy as an adaptive cell stress response mechanism. F1000Res. 2017; 6:2134.

145. Schonthal AH. Endoplasmic reticulum stress: its role in disease and novel prospects for therapy. Scientifica (Cairo). 2012; 2012:857516.
146. Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014; 14:581–97.

147. Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012; 13:89–102.
148. Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002; 16:452–66.

149. Maurel M, Chevet E, Tavernier J, Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci. 2014; 39:245–54.

150. Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013; 15:481–90.
151. Bartkowiak K, Kwiatkowski M, Buck F, Gorges TM, Nilse L, Assmann V, et al. Disseminated tumor cells persist in the bone marrow of breast cancer patients through sustained activation of the unfolded protein response. Cancer Res. 2015; 75:5367–77.

152. Pommier A, Anaparthy N, Memos N, Kelley ZL, Gouronnec A, Yan R, et al. Unresolved endoplasmic reticulum stress engenders immune-resistant, latent pancreatic cancer metastases. Science. 2018; 360:eaao4908.

153. Ranganathan AC, Zhang L, Adam AP, Aguirre-Ghiso JA. Functional coupling of p38-induced up-regulation of BiP and activation of RNA-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res. 2006; 66:1702–11.

154. Jaud M, Philippe C, Di Bella D, Tang W, Pyronnet S, Laurell H, et al. Translational regulations in response to endoplasmic reticulum stress in cancers. Cells. 2020; 9:540.

155. Kehrl JH, Sinnarajah S. RGS2: a multifunctional regulator of G-protein signaling. Int J Biochem Cell Biol. 2002; 34:432–8.

156. Nguyen CH, Ming H, Zhao P, Hugendubler L, Gros R, Kimball SR, et al. Translational control by RGS2. J Cell Biol. 2009; 186:755–65.

157. Zmijewski JW, Song L, Harkins L, Cobbs CS, Jope RS. Oxidative stress and heat shock stimulate RGS2 expression in 1321N1 astrocytoma cells. Arch Biochem Biophys. 2001; 392:192–6.
158. Ota A, Sawai M, Sakurai H. Stress-induced transcription of regulator of G protein signaling 2 (RGS2) by heat shock transcription factor HSF1. Biochimie. 2013; 95:1432–6.

159. Nguyen CH, Zhao P, Sobiesiak AJ, Chidiac P. RGS2 is a component of the cellular stress response. Biochem Biophys Res Commun. 2012; 426:129–34.

160. Park SY, Nam JS. The force awakens: metastatic dormant cancer cells. Exp Mol Med. 2020; 52:569–81.

161. Adamski V, Hattermann K, Kubelt C, Cohrs G, Lucius R, Synowitz M, et al. Entry and exit of chemotherapeutically-promoted cellular dormancy in glioblastoma cells is differentially affected by the chemokines CXCL12, CXCL16, and CX3CL1. Oncogene. 2020; 39:4421–35.
162. Hirano R, Okamoto K, Shinke M, Sato M, Watanabe S, Watanabe H, et al. Tissue-resident macrophages are major tumor-associated macrophage resources, contributing to early TNBC development, recurrence, and metastases. Commun Biol. 2023; 6:144.

163. Liu H, Ling CC, Yeung WH, Pang L, Liu J, Zhou J, et al. Monocytic MDSC mobilization promotes tumor recurrence after liver transplantation via CXCL10/TLR4/MMP14 signaling. Cell Death Dis. 2021; 12:489.

164. Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science. 2018; 361:eaao4227.

165. Hast J, Schiffer IB, Neugebauer B, Teichman E, Schreiber W, Brieger J, et al. Angiogenesis and fibroblast proliferation precede formation of recurrent tumors after radiation therapy in nude mice. Anticancer Res. 2002; 22:677–88.
166. Cho J, Min HY, Pei H, Wei X, Sim JY, Park SH, et al. The ATF6-EGF pathway mediates the awakening of slow-cycling chemoresistant cells and tumor recurrence by stimulating tumor angiogenesis. Cancers (Basel). 2020; 12:1772.

167. Huang X, Wang L, Guo H, Zhang W, Shao Z. Single-cell transcriptomics reveals the regulative roles of cancer associated fibroblasts in tumor immune microenvironment of recurrent osteosarcoma. Theranostics. 2022; 12:5877–87.
168. Bushnell GG, Deshmukh AP, den Hollander P, Luo M, Soundararajan R, Jia D, et al. Breast cancer dormancy: need for clinically relevant models to address current gaps in knowledge. NPJ Breast Cancer. 2021; 7:66.

169. Heidrich I, Deitert B, Werner S, Pantel K. Liquid biopsy for monitoring of tumor dormancy and early detection of disease recurrence in solid tumors. Cancer Metastasis Rev. 2023; 42:161–82.

170. Morin C, Moyret-Lalle C, Mertani HC, Diaz JJ, Marcel V. Heterogeneity and dynamic of EMT through the plasticity of ribosome and mRNA translation. Biochim Biophys Acta Rev Cancer. 2022; 1877:188718.

171. Brown MS, Abdollahi B, Wilkins OM, Lu H, Chakraborty P, Ognjenovic NB, et al. Phenotypic heterogeneity driven by plasticity of the intermediate EMT state governs disease progression and metastasis in breast cancer. Sci Adv. 2022; 8:eabj8002.

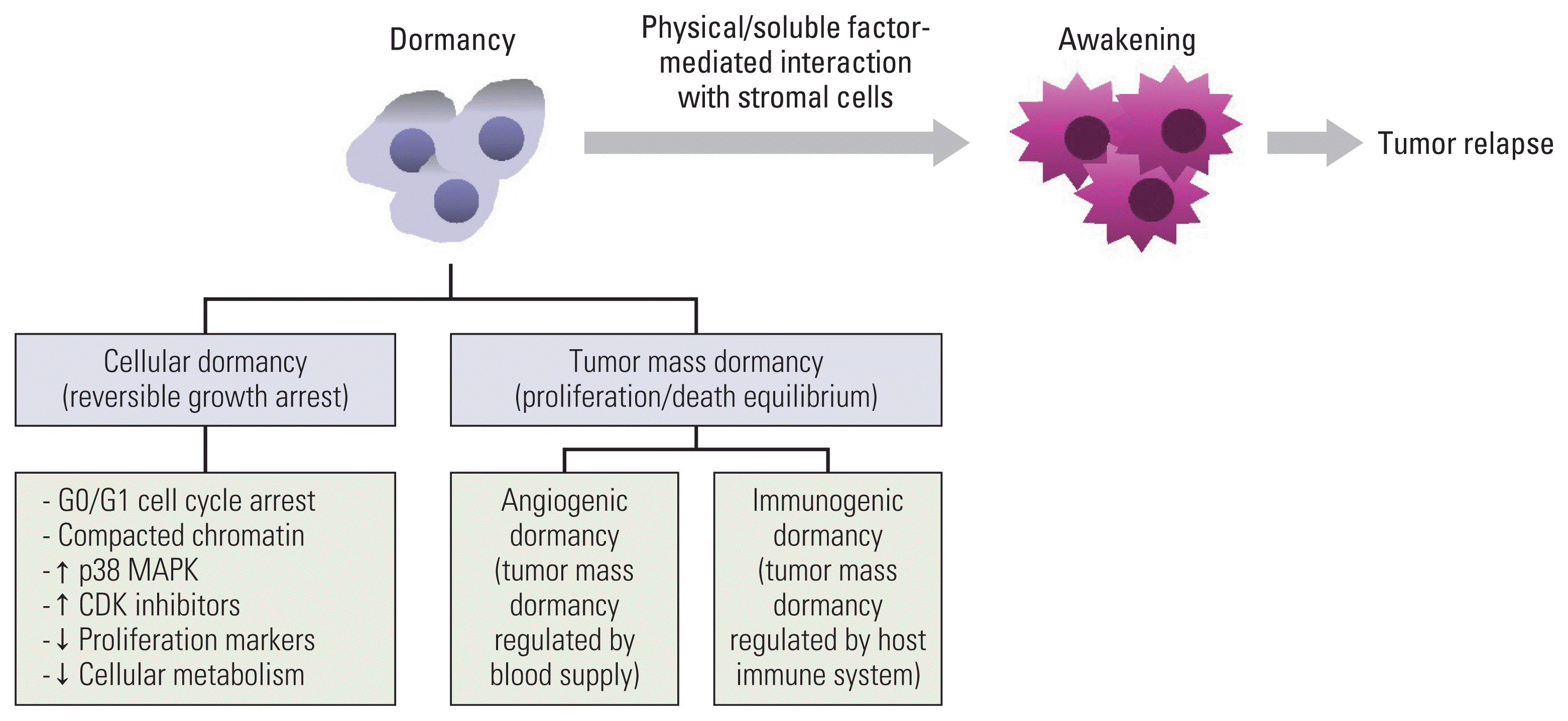
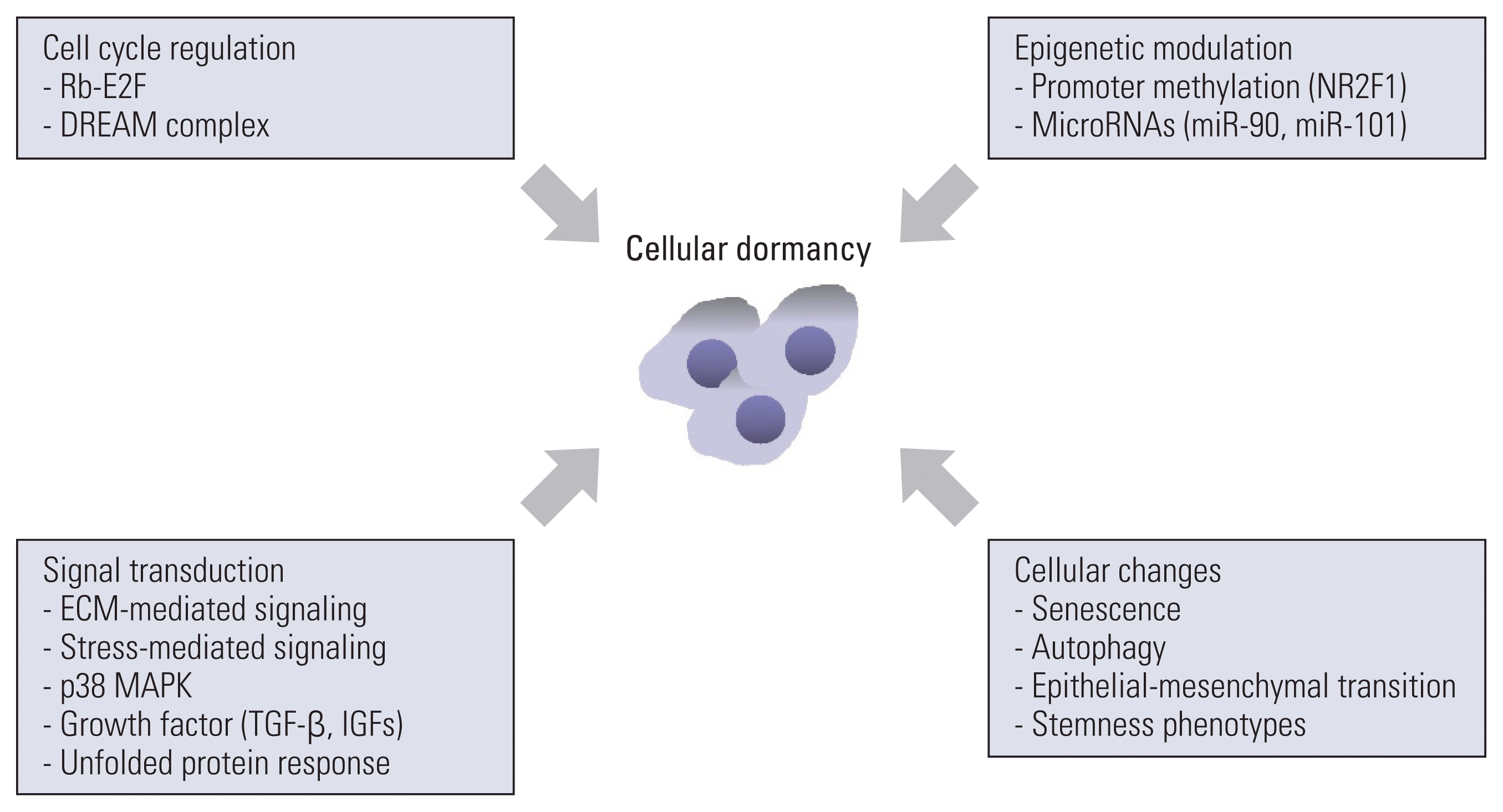
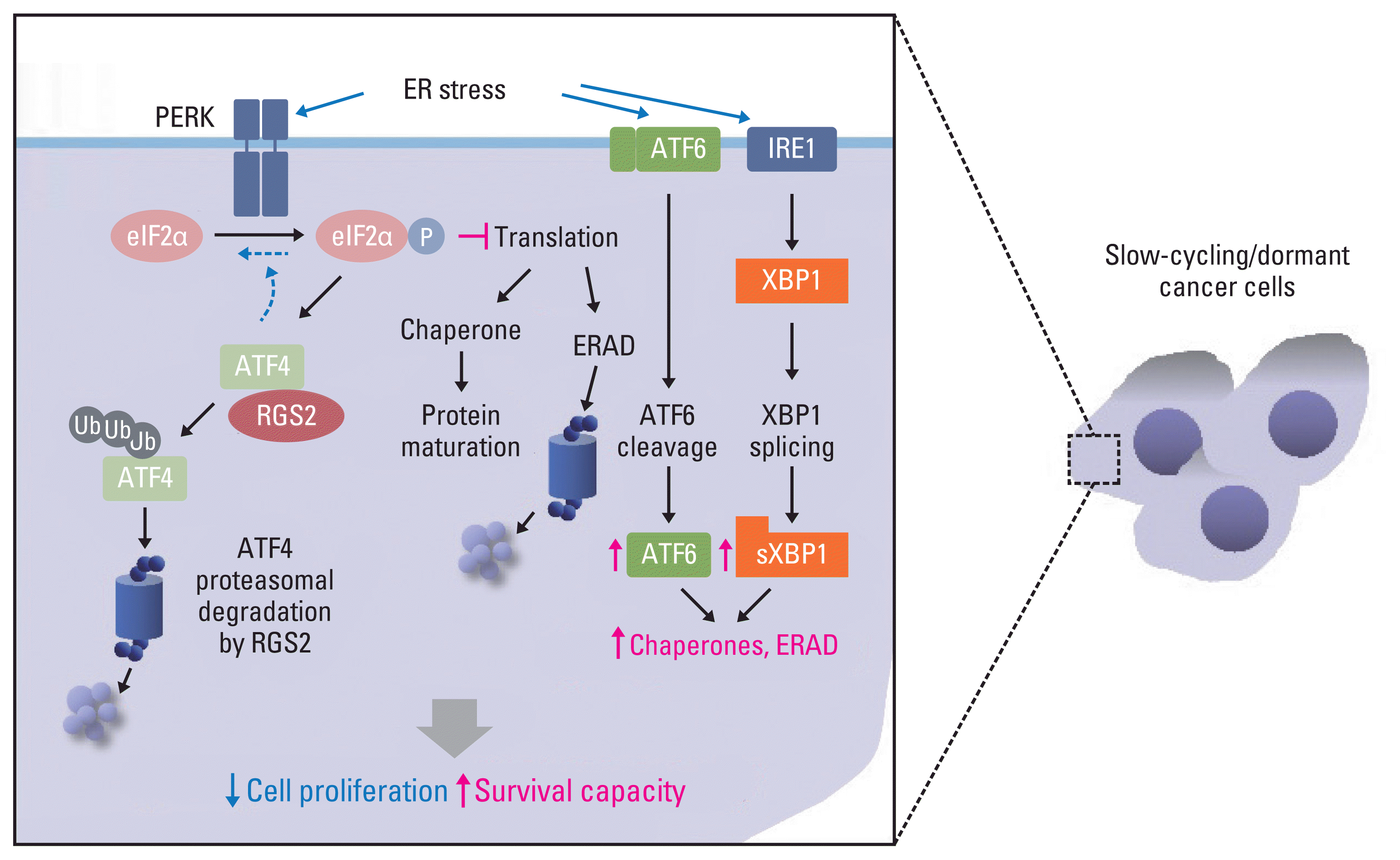
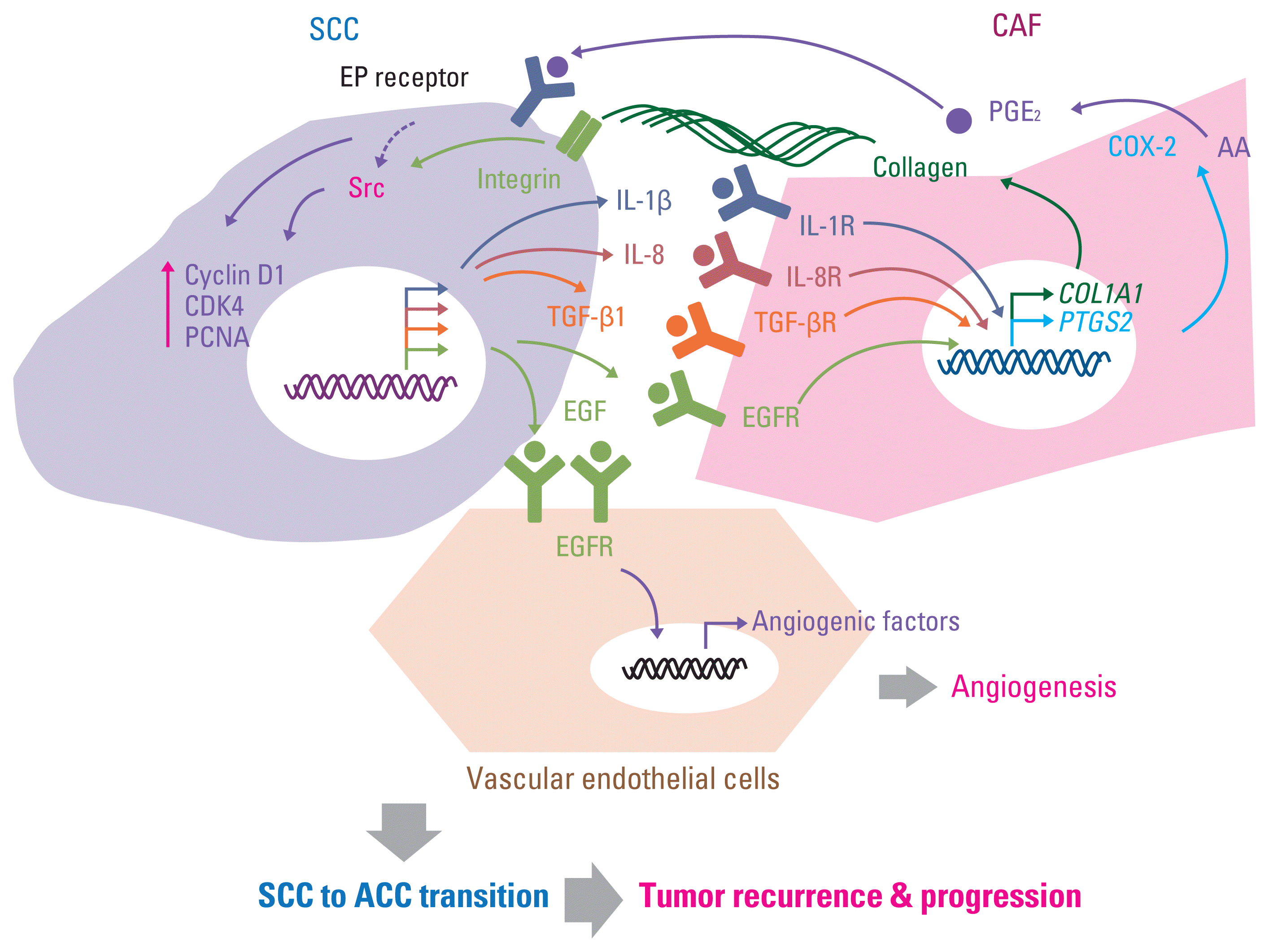




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download