Abstract
Purpose
Materials and Methods
Results
Notes
Ethical Statement
The study was approved by the Institutional Review Board (IRB) of Chonnam National University Hwasun Hospital, Korea (IRB reference number: CNUHH-2014-153). All study participants provided written informed consent.
Author Contributions
Conceived and designed the analysis: Ahn SY, Kim HJ (Hyeoung-Joon Kim), Ahn JS, Kim DDH.
Collected the data: Ahn SY, Kim M, Song GY, Jung SH, Yang DH, Lee JJ, Kim MY, Jung CW, Jang JH, Kim HJ (Hee Je Kim), Moon JH, Sohn SK, Won JH, Kim SH, Ahn JS, Kim DDH.
Contributed data or analysis tools: Ahn SY, Kim T, Ahn JS, Kim DDH.
Performed the analysis: Ahn SY, Kim T, Ahn JS.
Wrote the paper: Ahn SY, Ahn JS.
Acknowledgments
References
Fig. 1
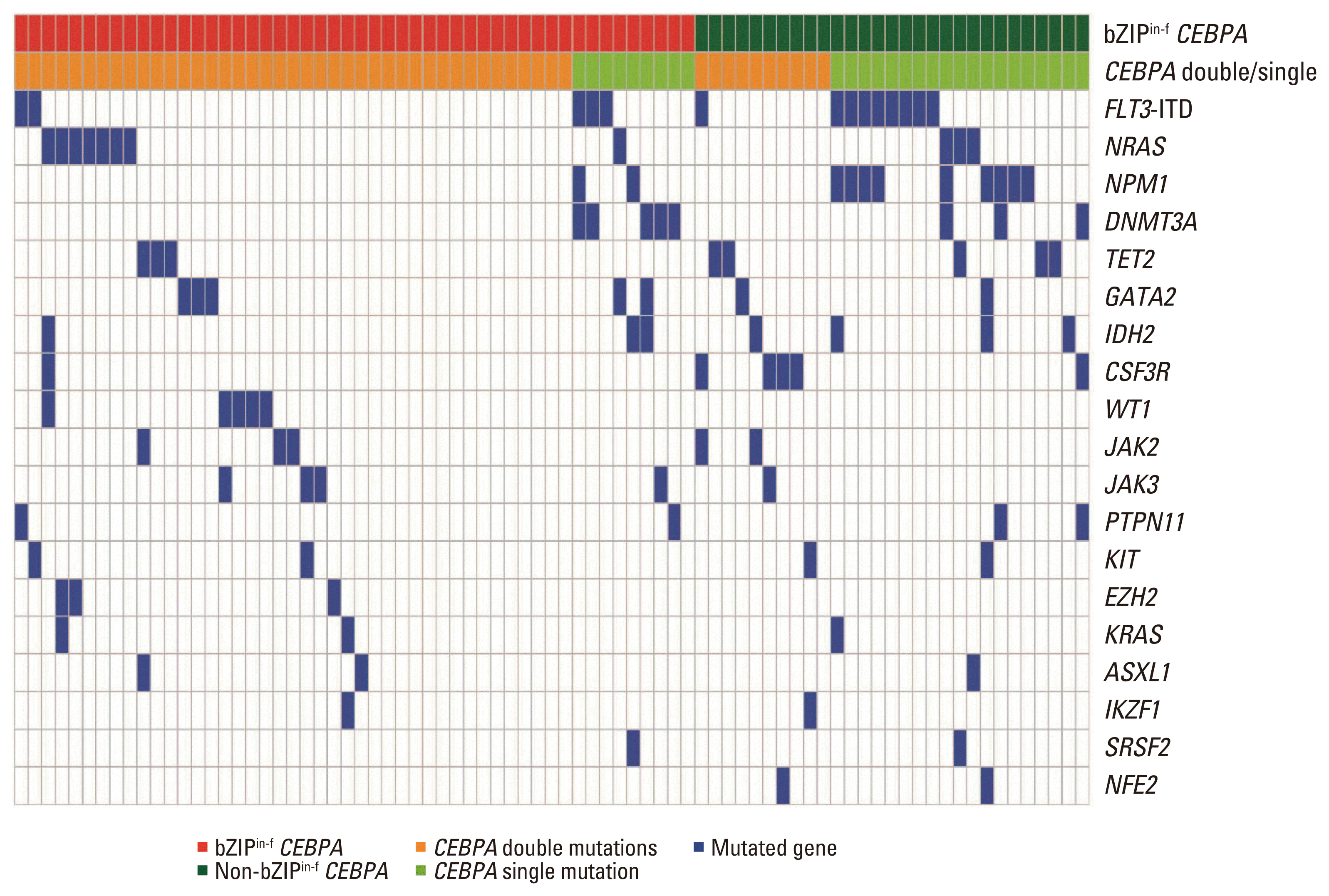
Fig. 2
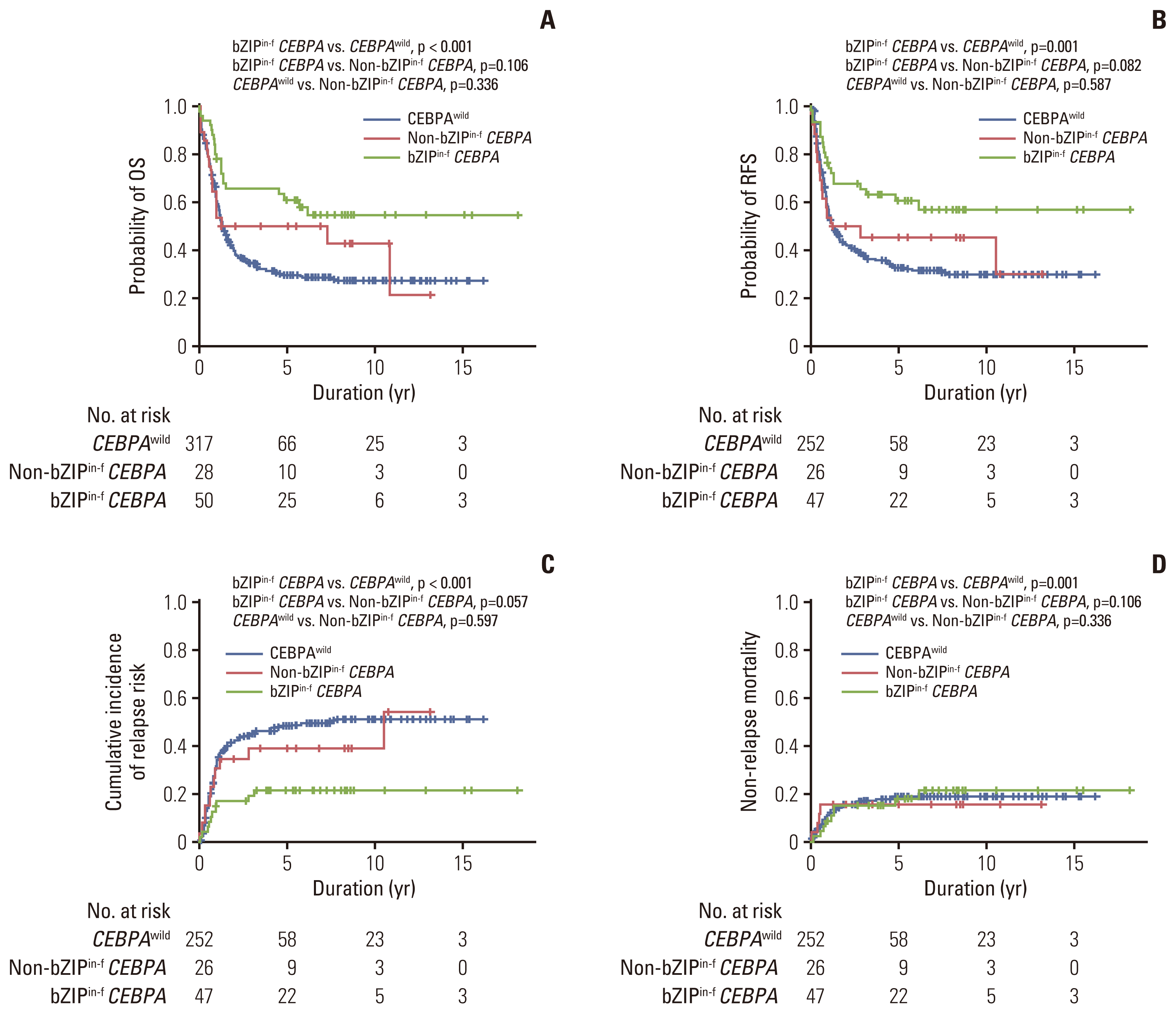
Fig. 3
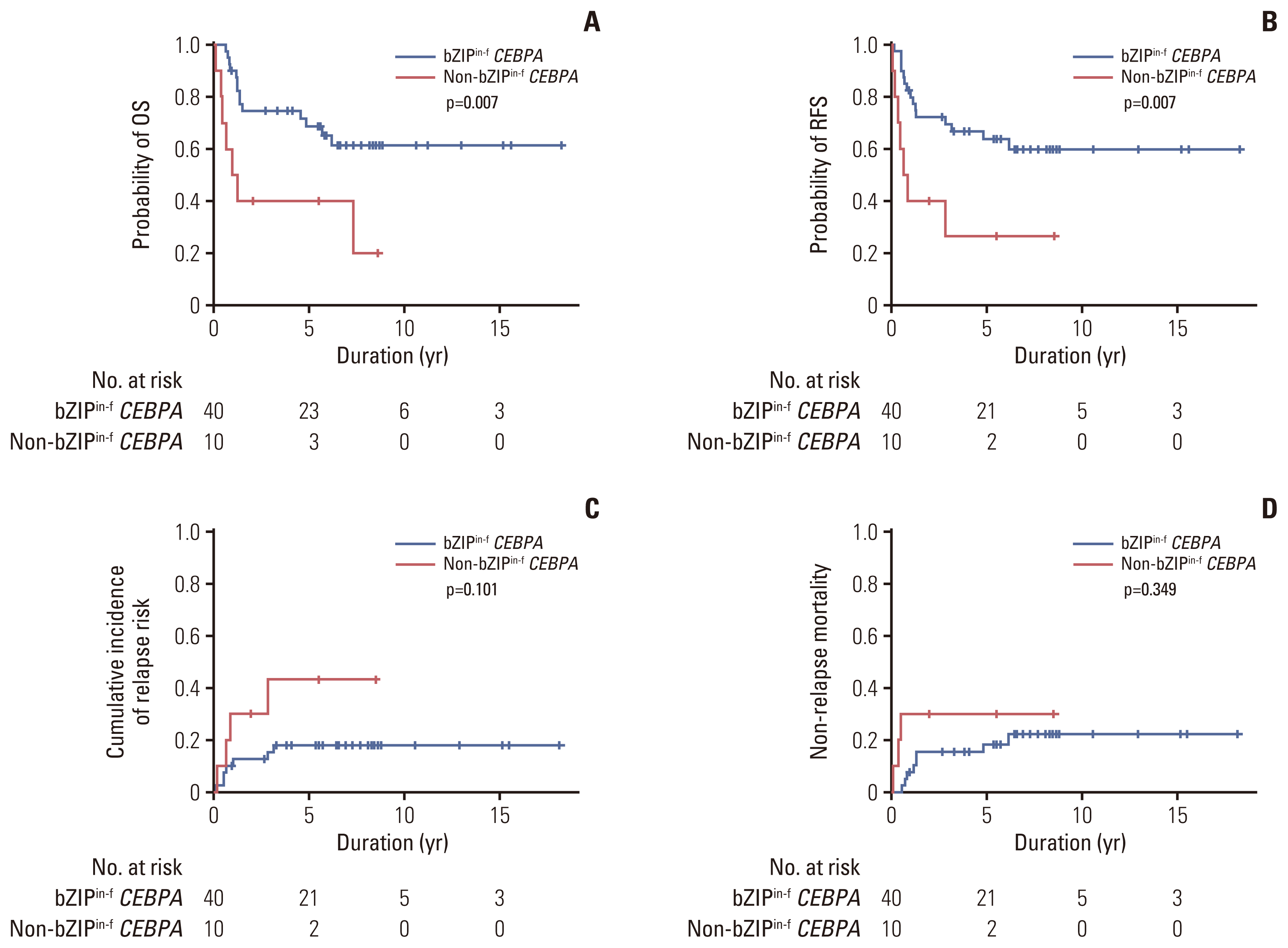
Fig. 4
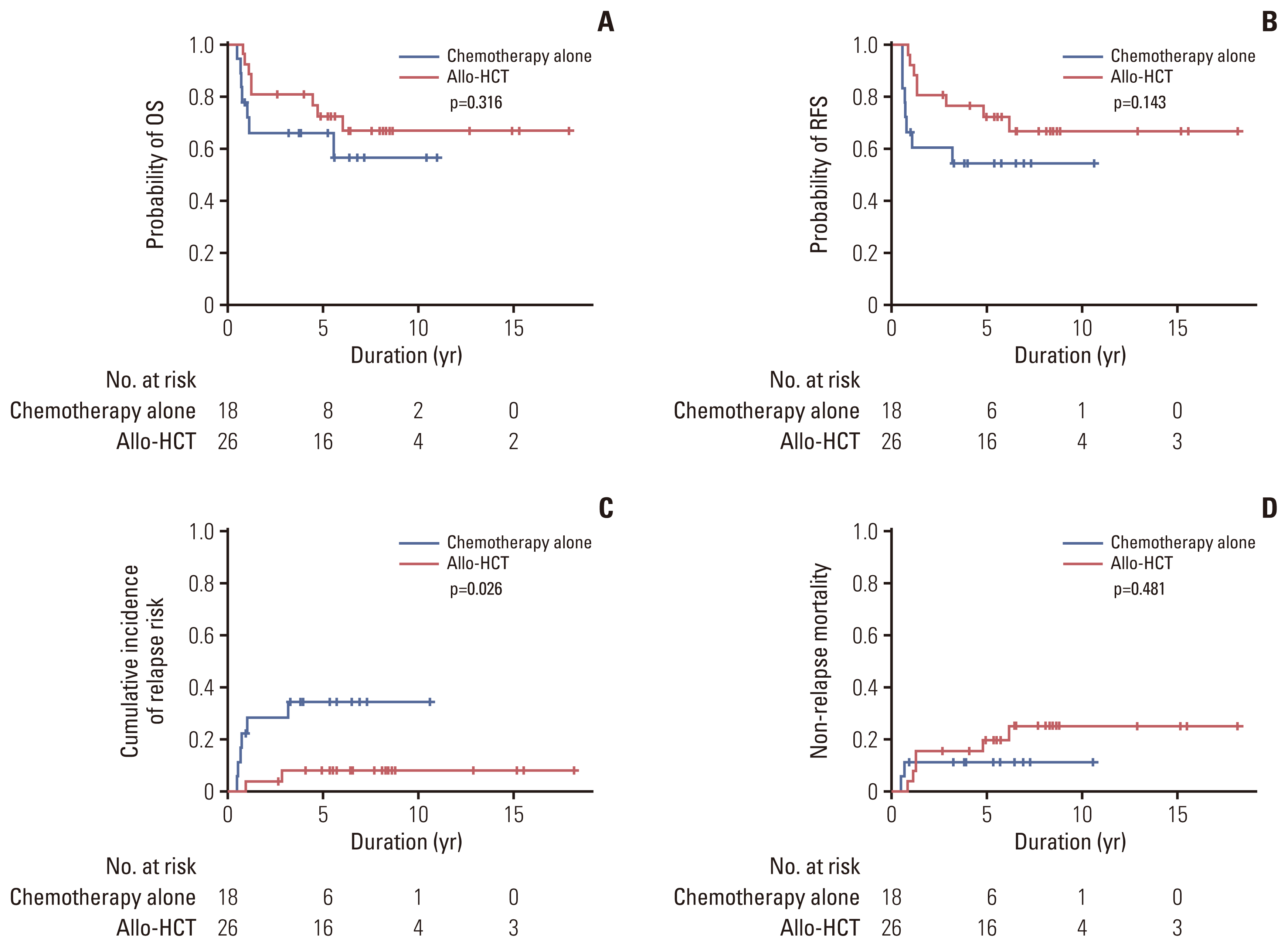
Fig. 5
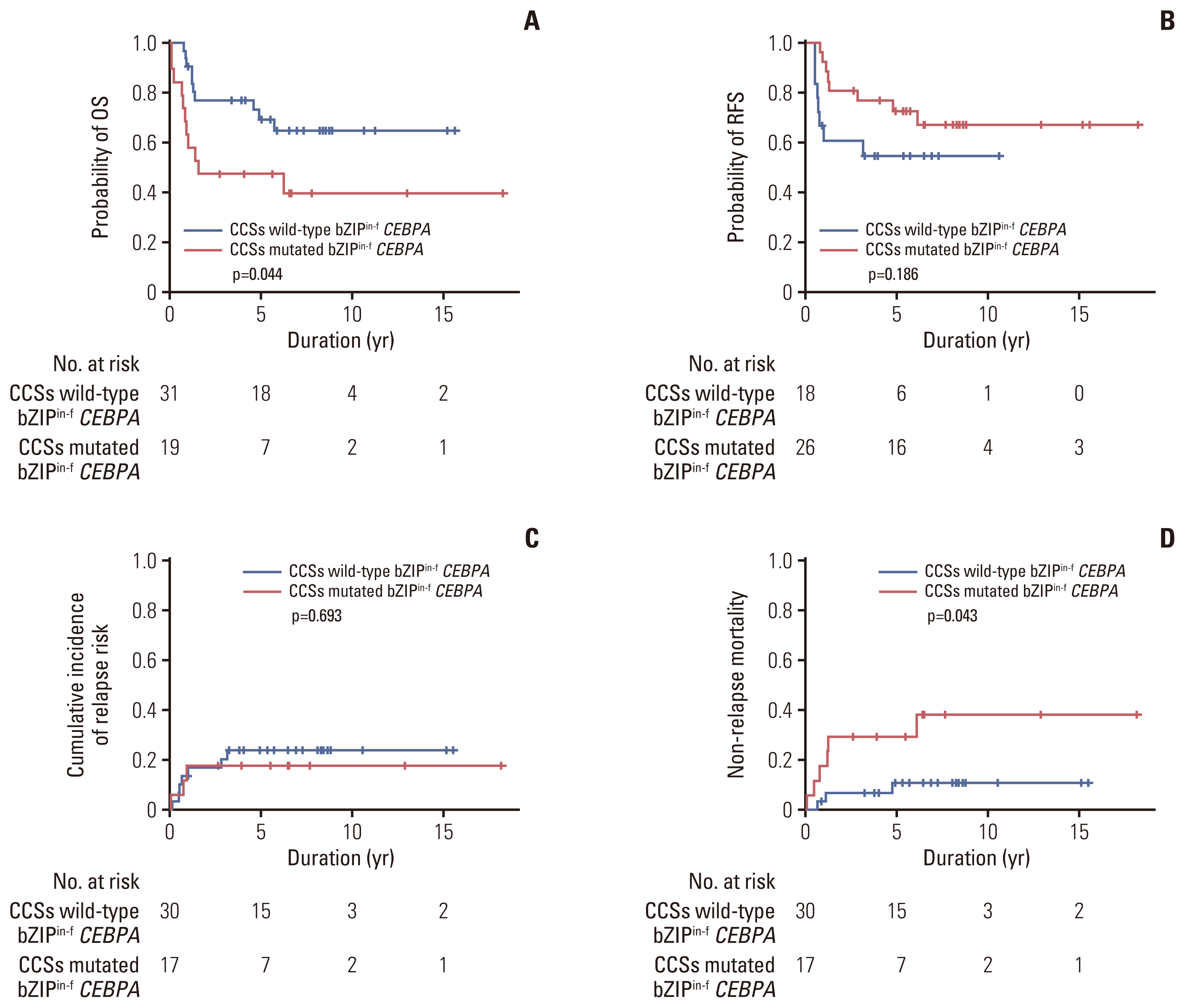
Table 1
Table 2
| Parameter | Median or frequency (n=395) | CEBPAwild (n=317) | bZIPin-f CEBPA mutations (n=50) | Non-bZIPin-f CEBPA mutations (n=28) | p-valuea) (wild vs. bZIPin-f) | p-valueb) (wild vs. non-bZIPin-f) | p-valuec) (bZIPin-f vs. non-bZIPin-f) |
|---|---|---|---|---|---|---|---|
| Age (yr) | 52 (15–84) | 54 (15–84) | 38 (15–67) | 57 (17–75) | < 0.001d) | 0.793 | 0.001d) |
| Male sex | 201 (50.9) | 161 (50.3) | 23 (63.0) | 17 (43.1) | 0.547 | 0.332 | 0.244 |
| WBC (×109/L) | 28.2 (0.3–397.2) | 27.1 (0.3–384.0) | 31.6 (2.0–333.2) | 49.8 (1.7–397.2) | 0.142 | 0.031d) | 0.327 |
| Bone marrow blasts (%) | 63 (3–100) | 70 (3–100) | 82 (21–100) | 70 (30–100) | 0.135 | 0.231 | 0.964 |
| NPM1mut | 128 (32.4) | 117 (36.9) | 2 (4.0) | 9 (32.1) | < 0.001d) | 0.686 | 0.001d) |
| FLT3-ITDmut | 107 (27.1) | 93 (29.3) | 5 (10.0) | 9 (32.1) | 0.003d) | 0.829 | 0.028d) |
| DNMT3Amut | 95 (24.1) | 87 (27.4) | 5 (10.0) | 3 (10.7) | 0.008d) | 0.070 | > 0.99 |
| NRASmut | 68 (17.2) | 57 (18.0) | 8 (16.0) | 3 (10.7) | 0.844 | 0.440 | 0.737 |
| IDH2mut | 55 (13.9) | 48 (15.1) | 3 (6.0) | 4 (14.3) | 0.120 | > 0.99 | 0.243 |
| PTPN11mut | 36 (9.1) | 32 (10.1) | 2 (4.0) | 2 (7.1) | 0.289 | > 0.99 | 0.615 |
| FLT3-TKDmut | 33 (8.4) | 32 (10.1) | 1 (2.0) | 0 | 0.065 | 0.092 | > 0.99 |
| IDH1mut | 32 (8.1) | 31 (9.8) | 0 | 1 (3.6) | 0.013d) | 0.494 | 0.359 |
| TET2mut | 27 (6.8) | 20 (6.3) | 3 (6.0) | 4 (14.3) | > 0.99 | 0.118 | 0.243 |
| KRASmut | 22 (5.6) | 19 (6.0) | 2 (4.0) | 1 (3.6) | 0.752 | > 0.99 | > 0.99 |
| SRSF2mut | 21 (5.3) | 19 (6.0) | 1 (2.0) | 1 (3.6) | 0.497 | > 0.99 | > 0.99 |
| GATA2mut | 16 (4.1) | 9 (2.8) | 5 (10.0) | 2 (7.1) | 0.029d) | 0.222 | > 0.99 |
| Achieved CR | 325 (82.3) | 252 (79.5) | 47 (94.0) | 26 (92.9) | 0.011d) | 0.131 | > 0.99 |
| Received allo-HCT in CR achieved patients (n=325) | 127 (39.1) | 91 (36.1) | 26 (52.0) | 10 (35.7) | 0.015d) | > 0.99 | 0.223 |
Values are presented as median (range) or number (%). Allo-HCT, allogeneic hematopoietic cell transplantation; bZIPin-f CEBPA, bZIP in-frame CEBPA mutation; CEBPA, CCAAT/enhancer-binding protein α; CEBPAwild, CEBPA wild-type; CR, complete remission; non-bZIPin-f CEBPA, non bZIP or non-in-frame CEBPA mutation; WBC, white blood cell.
Table 3
Allo-HCT, allogeneic hematopoietic cell transplantation; bZIPin-f CEBPA, bZIP in-frame CEBPA mutation; CEBPA, CCAAT/enhancer-binding protein α; CI, confidence internal; CIR, cumulate incidence of relapse; CR, complete remission; HR, hazard ratio; OR, odd ratio; OS, overall survival; RFS, relapse-free survival.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download