INTRODUCTION
Canine parvovirus (CPV) causes acute hemorrhagic gastroenteritis, nausea, diarrhea, leukopenia, and lethal myocarditis in puppies. It is a small, non-enveloped, single-stranded linear DNA virus that is 18~26 nm in diameter (1). It belongs to the genus Protoparvovirus, family Parvoviridae subfamily Parvovirinae, and is closely related to feline panleukopenia virus (FPV). The CPV genome is approximately 5.2 kb in length and encodes two non-structural (NS1 and NS2) and two structural (VP1 and VP2) proteins (2). The VP2 capsid protein is the major protein determining antigenicity and host range and induces protective neutralizing antibodies (3). Fecal–oral CPV transmission has been reported in domestic dogs and wild carnivores (4).
The CPV genotypes are divided into CPV-2, CPV-2a, CPV-2b, and CPV-2c based on amino acid substitutions in the VP2 gene. CPV-2 emerged in the late 1970s and became widespread in Europe, Asia, and North America (5). In 1985, CPV-2 was replaced by the variants CPV-2a and CPV-2b (6). CPV-2a has substitutions in the amino acid sequence of the VP2 gene at positions Met87Leu, Gly300Ala, Tyr305Asp, and Val 555Ile compared to the original CPV-2, while CPV-2b differs from CPV-2a at Asn426Asp and Ile555Val (7). A new CPV-2a/CPV-2b was reported to have an amino acid mutation at position Ser297Ala and appears to have replaced the original CPV-2a/2b (8). CPV-2c with glutamic acid at position 426 of the VP2 protein was reported in Italy in 2000 (9). Recently, CPV-2c has been circulating in Spain, Japan, Taiwan, and Uruguay (10, 11). The antigenic variants named New CPV-2a and New CPV-2b have been continuously replacing the CPV-2 variants.
Attenuated CPV vaccines to reduce parvovirus disease in puppies were developed using the CPV-2 strain although they vary depending on the manufacturer (12). However, CPV-2 variants have emerged rapidly in the canine population and these variants may reduce the effectiveness of CPV-2-based vaccines at protecting against parvoviral disease (13). Thus, CPV genotyping of new CPV infections is necessary to understand the evolution of the virus and provide information about its field status. In this study, we isolated two wild CPV-2a isolates circulating in a Korean dog population and determined their biological and molecular features based on sequence analysis of the VP2 gene.
MATERIALS AND METHODS
Cell and virus isolation
A72 cells (ATCC, CRL-1542) derived from canine fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), two antibiotics (penicillin and streptomycin), and an antimycotic (amphotericin B). Two feces samples were obtained from diseased dogs in Gimcheon, South Korea, in 2021. For virus isolation, the samples were filtered through a 0.45 µm syringe filter and inoculated into A72 cells. The inoculated cells were maintained at 37°C in a humidified 5% CO2 incubator, and their cytopathic effect (CPE) was observed for 7 days. The two isolates were designated CPV2101 and CPV2102.
Immunofluorescence assay (IFA)
To confirm viral proliferation in A72 cells, A72 cells were infected with each isolate and fixed in cold 80% acetone. The fixed cells were incubated at 37°C with a specific CPV monoclonal antibody (MEDIAN Diagnostics, Chuncheon, Korea) and stained with fluorescein isothiocyanate-conjugated goat anti-mouse IgG + IgM (KPL Laboratories, USA). After washing in phosphate-buffered saline (pH 7.2), specific fluorescence was observed in the nuclei of the infected cells using a fluorescence microscope (Nikon, Tokyo, Japan).
Hemagglutination assay (HA)
Pig erythrocytes were washed three times with Sorensen buffer (pH 6.8) and prepared in the same buffer at a concentration of 0.7% containing 0.17% bovine serum albumin. First, 25 μL each isolate was diluted two-fold in Sorensen buffer in U-96-well plates. Then, 25 μL washed RBCs were added to each well, and the plates were incubated at 4°C for 1 h. The HA titer was expressed as the reciprocal of the highest dilution of the isolate showing hemagglutination.
Electron microscopy (EM)
To determine the morphology and particle size of the two isolates, virus supernatants grown in A72 cells were treated with 10% polyethylene glycol (MW 8000; Sigma-Aldrich, Missouri, USA) and 0.5 M NaCl and precipitated at 4°C overnight. The supernatants were purified by ultracentrifugation with a cesium chloride (CsCl) gradient at 32,000 rpm at 4°C. The purified virus bands were located between the densities of 1.3 and 1.5 g/cm3 in CsCl. The purified virus was stained negatively with 1% uranyl acetate and subjected to transmission electron microscopy (Hitachi 7100; Hitachi, Tokyo, Japan).
Real-time PCR
Real-time PCR was carried out on each isolate. The primers 5'ACA CCT GAG AGA TTT ACA TAT ATA GCA CA3' (F) and 5'ATT AGT ATA GTT AAT TCC TGT TTT ACC TCC3' (R) were designed specifically for the CPV VP2 gene and gave a 159 bp product. PCR was performed in a real-time PCR thermal cycler (qTOWER3, Analytic Jena, Jena, Germany) in 20 μL reaction volumes containing 2 μL DNA, 10 pmol each primer, 10 μL SYBR Green Master mix (Roche, Germany), and sterile water. The PCR consisted of 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 60°C for 10 s, 72°C for 15 s, and a final 10 min extension at 72°C.
Cell tropism
The host cell tropism of each isolate was assessed using A72, CRFK (ATCC, CCL-94), BHK-21 (ATCC, CCL-10), MDCK (ATCC, CCL-34), Vero (ATCC, CCL-81), and MDBK (ATCC, CCL-22) cells derived from canine fibroblasts, feline kidney cells, hamster fibroblasts, canine kidney cells, simian kidney cells, and bovine kidney cells, respectively. The six kinds of cells were cultured in DMEM supplemented with 10% FBS. The cell monolayers were washed twice with PBS and inoculated with each isolate. Following adsorption, the culture fluid was replaced with new DMEM containing 3% FBS. The viral supernatants were harvested after 4 days and viral proliferation was examined via IFA, as described above.
DNA extraction and whole genome sequencing
The complete genome of each isolate was sequenced using a MiSeq (Illumina, San Diego, USA) next-generation sequencing (NGS) platform. Briefly, DNA was extracted from the viral supernatant using a DNA extraction kit (QIAGEN, Valentia, USA) and sequenced by Sanigen (Anyang, South Korea) using the TruSeq Nano DNA prep kit (Illumina). Contigs were obtained, and open reading frames (ORFs) were predicted by the National Center for Biotechnology Information (NCBI) ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). A BLASTn search of the NCBI database for the sequences was performed. Default parameters were used for all software unless otherwise specified.
Sequence analyses
The amino acid sequences of the VP2 gene (1,755 nt, 584 aa) were aligned using ClustalW and compared to the VP2 sequences of reference strains (FPV, CPV-2, CPV-2a, New CPV-2a, CPV-2b, New CPV-2b, and CPV-2c) retrieved from GenBank. Then these alignments were subjected to deduced amino acid analysis with BioEdit. For homology analysis of the VP2 sequence, Bayesian phylogenetic analyses were performed based on the full-length VP2 nucleotide sequences of the two isolates. A phylogenetic tree was constructed using Bayesian Markov Chain Monte Carlo (MCMC) analysis implemented in Bayesian evolutionary analysis sampling trees (BEAST) v1.10.4 (14).
RESULTS
Virus isolation and identification
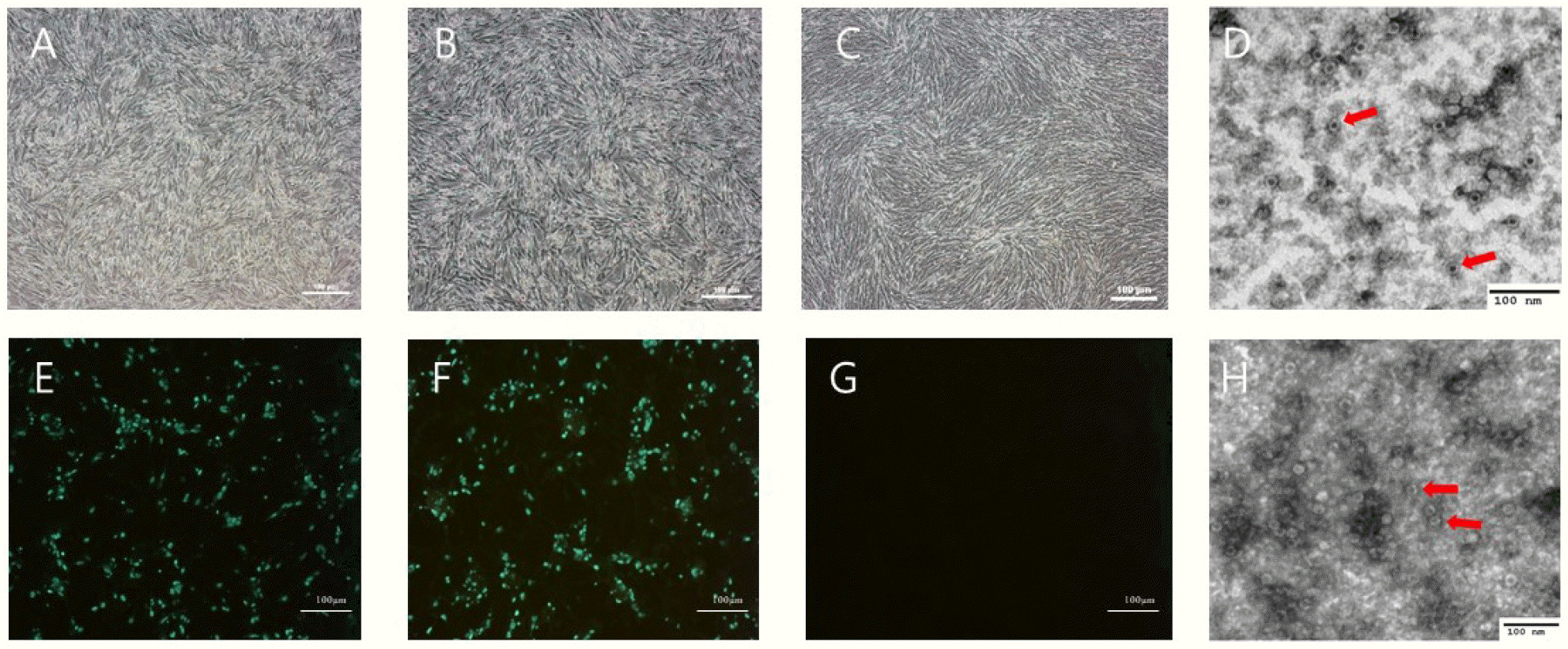
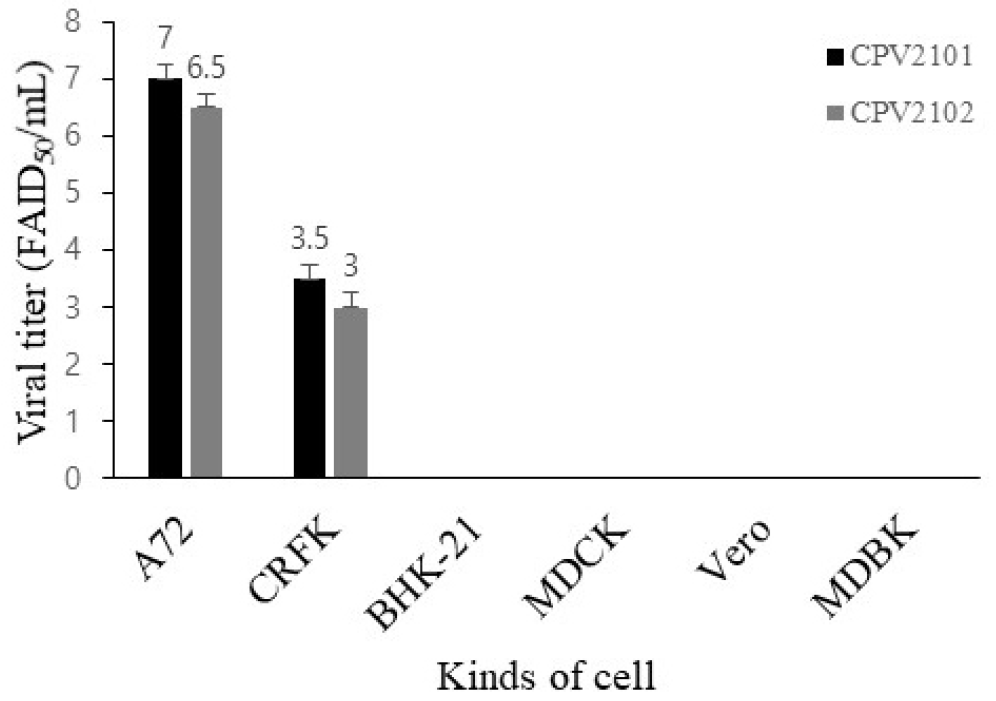
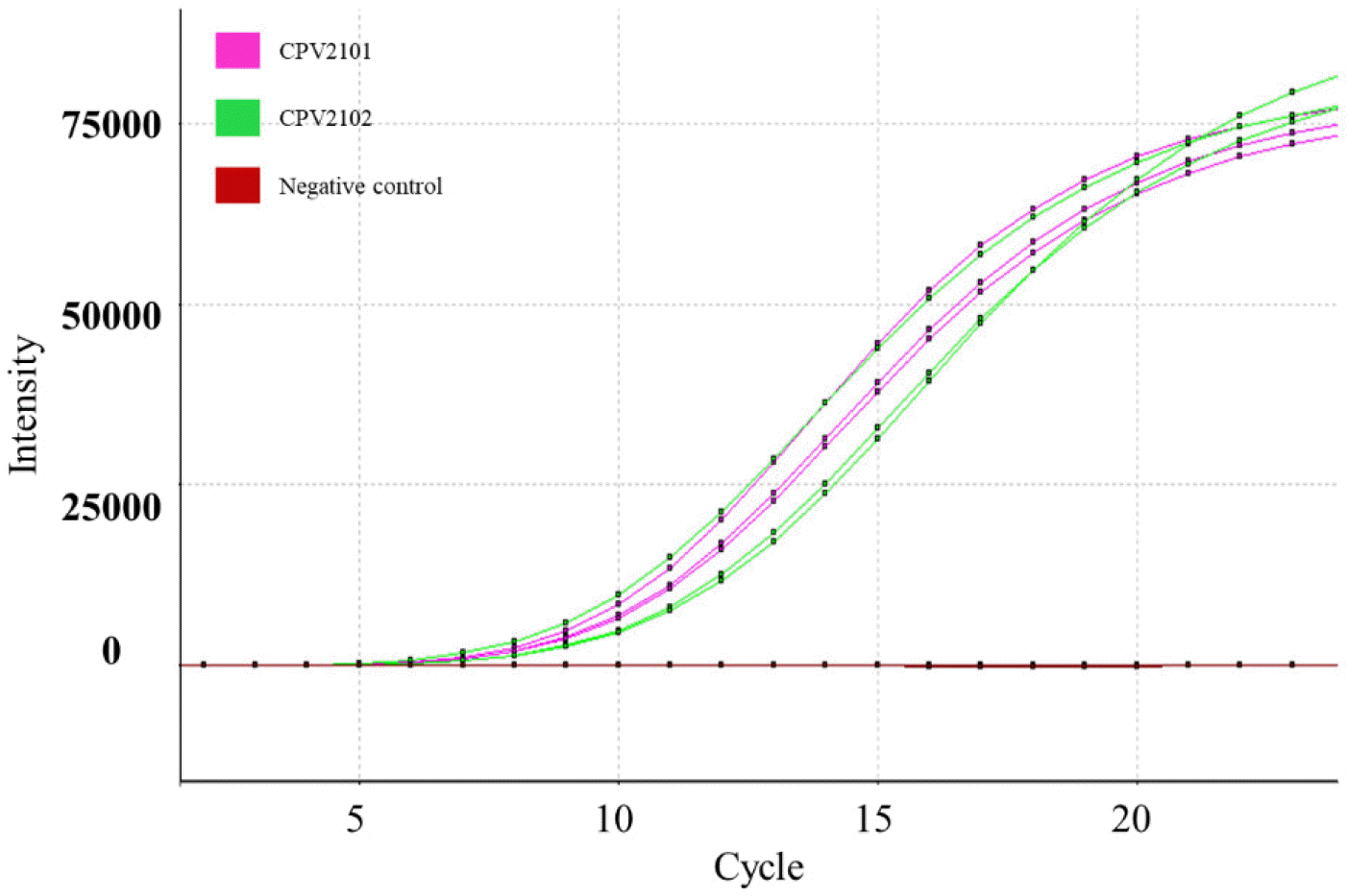
Six days after inoculating filtered feces samples into A72 cells, CPE was observed under a microscope (Fig. 1). The two isolates had HA titers ranging from 211 to 212. To confirm virus identification in A72 cells, A72 cells infected with the isolates were stained with CPV-specific monoclonal antibodies. Specific fluorescence was observed in the nuclei of the infected A72 cells. To determine the morphology and size of the isolates, the isolates were concentrated and purified. A typical parvovirus particle measuring approximately 23 nm in diameter was observed by electron microscopy. The isolate titers ranged from 107.0 to 106.5 FAID50/mL in A72 cells and 103.5 to 103.0 FAID50/mL in CRFK cells (Fig. 2), indicating that both isolates were capable of infecting CRFK and A72 cells, but not cells originating from other species. The 159 bp VP2 gene fragment amplified from the DNA extracted from the two isolates by real-time PCR was seen the CRFK and A72 cells (Fig. 3).
Fig. 1
CPV2101 and CPV2102 strains isolated from A72 cells. Appearance of the cytopathic effect in A72 cells infected with isolates CPV2101 (A) and CPV2102 (B) and normal A72 cells (C, G). Immunofluorescence assay of A72 cells infected with CPV2101 (E) and CPV2102 (F) using CPV-specific monoclonal antibody. Transmission electron micrograph images of the purified CPV2101 (D) and CPV2102 (H) isolates. Red arrows point to viral particles. Scale bars in A72 cells indicate 100 μm.
Analysis of the nucleotide and amino acid sequences
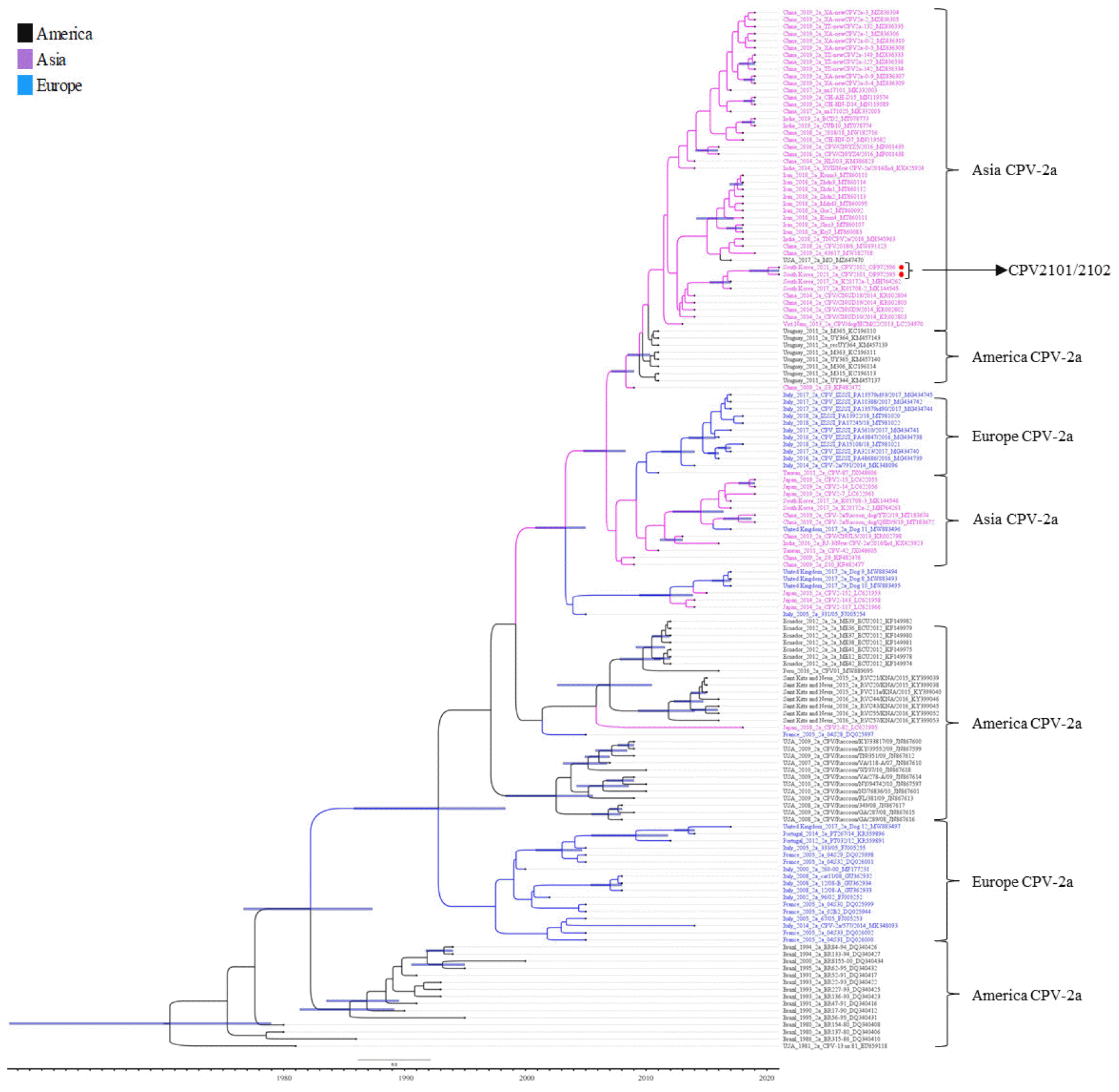
Genomic DNA of both isolates was subject to whole genome sequencing. The complete linear genomes of two isolates were assembled into single contigs with lengths of 5,032 and 5,001 bp (both 36.9% G+C content). The sequences of the two isolates were submitted to GenBank under accession numbers OP972595 and OP972596. The genomes of the two isolates had 100.0% nucleotide similarity to each other. Searching against the NCBI nucleotide database, the sequences of the two isolates had the closest nucleotide similarity (99.7%) to the Canine/China/24/2017 strain. The VP2 amino acid sequences of both isolates were analyzed with representative FPV, CPV-2, CPV-2a, New CPV-2a, CPV-2b, New CPV-2b, and CPV-2c reference sequences retrieved from GenBank. Both isolates were classified as CPV-2a with the amino acid substitutions Phe267Tyr, Thr322Ala, Tyr324Ile, and Thr440Ala (Table 1). In a phylogenetic tree constructed based on VP2 nucleotide sequences, both isolates clustered with Asia CPV-2a (Fig. 4). The phylogenetic analysis indicated that both isolates were closely related to K01708-1 but in separate subclusters from the American and European CPV-2a.
Table 1.
Differences in amino acid sequences of VP2 genes of eight canine parvoviruses and one feline panleukopenia virus strain
| Genotype isolates | Amino acid and positions of VP2 gene | Remark | Accession | |||||
|---|---|---|---|---|---|---|---|---|
| 267 | 297 | 322 | 324 | 426 | 440 | variants | numbers | |
| *FPV | Phe | Ser | Thr | Tyr | Asn | Thr | M38246 | |
| **CPV-2 | Phe | Ser | Thr | Tyr | Asn | Thr | M38245 | |
| CPV-2a | Phe | Ser | Thr | Tyr | Asn | Thr | D26079 | |
| CPV-2b | Phe | Ala | Thr | Tyr | Asp | Thr | EU659120 | |
| New CPV-2a | Phe | Ala | Thr | Tyr | Asn | Thr | AY742953 | |
| New CPV-2b | Phe | Ala | Thr | Tyr | Asp | Thr | AY742955 | |
| CPV-2c | Phe | Ala | Thr | Tyr | Glu | Thr | MF177239 | |
| CPV2101 | Tyr† | Ala | Ala | Ile† | Asn | Ala† | New CPV-2a | OP972595 |
| CPV2102 | Tyr | Ala | Ala | Ile | Asn | Ala | New CPV-2a | OP972596 |
Fig. 4
Phylogenetic and temporal relationship analysis of 147 CPV-2a strains based on the full VP2 gene (1,755 nucleotides). Red circles show CPV2101 and CPV2102. Node bars in blue indicate 95% highest posterior density. CPV-2a strains of Asian origin are shown in purple, of American origin in black, and of European origin in blue.
DISCUSSION
Since CPV was first identified, many variants have been described, some of which result in antigenic changes (13). Based on amino acid analysis of the VP2 gene, CPV-2 has been divided into genotypes CPV-2/2a/2b/2c, of which CPV-2a is the dominant genotype in Korean puppies (15, 16). In this study, two viruses were isolated from dogs in Gimcheon, South Korea, and it was confirmed that both were CPV using IFA, EM, and real-time PCR. The two isolates were classified as CPV-2a, indicating that CPV-2a is still circulating in the Korean dog population. Investigating the cell tropism of the isolates, both proliferated in A72 and CRFK cells and had titers of 107.0 and 103.5 FAID50/mL, respectively. This suggests that Korean CPV-2a causes infection in both dogs and cats. This needs to be verified in further studies by infecting cats with the CPV2101 and CPV2102 strains.
Analysis of the VP2 gene has confirmed the emergence of new CPV variants. The VP2 capsid protein determines its antigenicity, hemagglutination, and host range (3). In the amino acid sequence of the VP2 gene of the two isolates reported here, non-synonymous changes were observed at positions 267, 322, 324, and 440. Except for residue 322, these mutations have already been reported in other countries, including China and Uruguay (16, 17). Functionally, residue 267 does not affect the antigenicity of CPV because this residue is not located in an exposed area (18); residues 324 and 93 of VP2 regulate its host range (12). Because residue 440 is exposed on the capsid surface, mutation of this residue might affect antigenic changes (19). Therefore, the CPV2101 and CPV2102 strains may have antigenic variation compared to old CPV-2a. Phylogenetic analysis of the CPV VP2 gene is performed to study evolutionary relationships and find genetic mutants among CPV strains. Our two isolates were closely related to K01708-1, which was isolated in South Korea in 2017, and were located in the Asian CPV-2a clade along with CPV-2a strains isolated from China, South Korea, Iran, and India. This suggests that Asian CPV-2a is constantly evolving.
In conclusion, we isolated two CPVs named CPV2101 and CPV2102 and confirmed that they were new CPV-2a variants. The amino acid analysis indicated that the two strains had an alanine at position 297 and asparagine at position 426; amino acid mutations were found at four positions in the VP2 gene compared to CPV-2a. However, few investigations have characterized the pathology of CPV-2a circulating in dog populations in South Korea. Therefore, further study is needed to determine whether the newly isolated CPV-2a strains are pathogenic in dogs and cats.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download