INTRODUCTION
MATERIALS AND METHODS
Study populations
Genotyping and quality controls
Genome-wide SNP-SNP interactions via BOLL and EDNRA genes
RESULTS
Fig. 1.
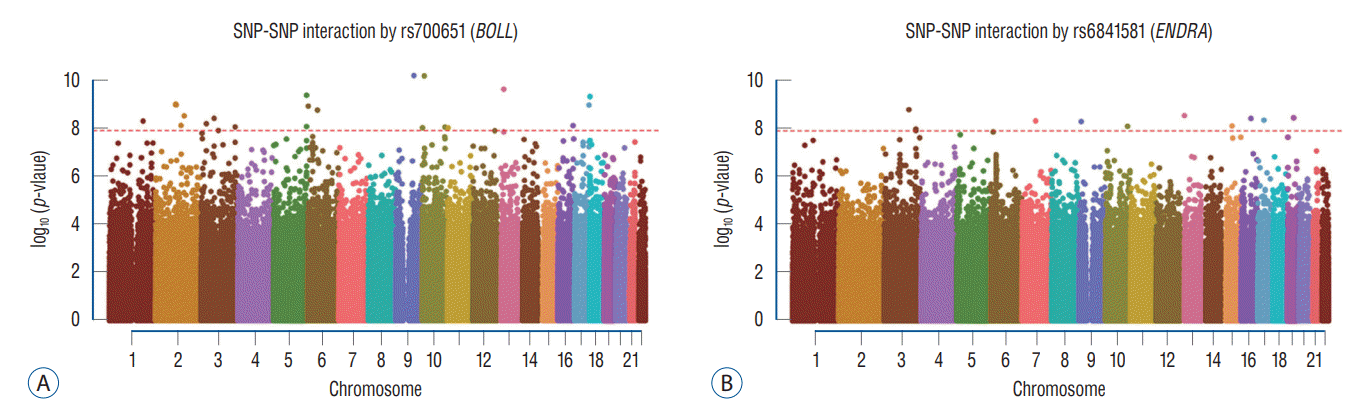
Fig. 2.
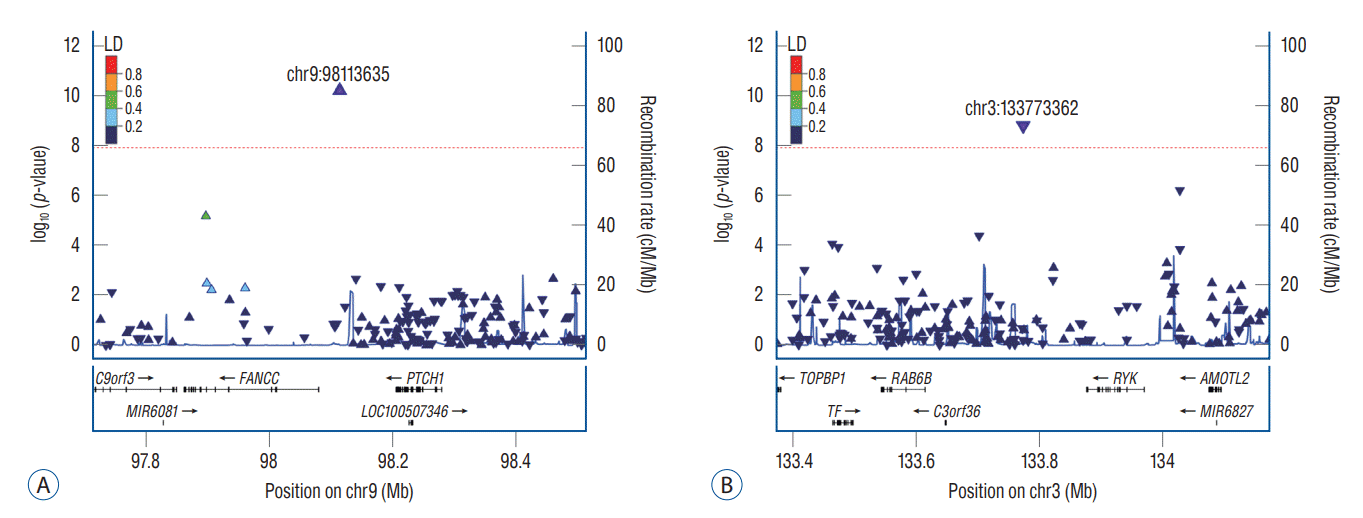
Table 1.
| Gene | Chr | SNP | BP | M/m | MAF, case/control | lnOR* | p-value for GWAS* | lnOR† | p-value for interaction† |
|---|---|---|---|---|---|---|---|---|---|
| BOLL | 2q33.1 | rs700651 | 198631714 | A/G | 0.476/0.449 | 1.42 | 0.0079 | NA | NA |
| PTCH1 | 9q22.32 | rs1105980 | 98113635 | G/C | 0.27/0.294 | -0.08 | 0.5821 | 1.53 | 6.41E-11 |
| CCDC3 | 10p13 | rs12412014 | 12911725 | G/C | 0.281/0.291 | -0.09 | 0.5145 | 1.47 | 6.63E-11 |
| LINC00457 | 13q13.2 | rs1536847 | 35106975 | G/T | 0.295/0.329 | -0.18 | 0.2033 | 1.39 | 2.37E-10 |
| C5orf60 | 5q35.3 | rs62405726 | 179069468 | G/A | 0.318/0.287 | 0.13 | 0.3646 | 1.46 | 4.21E-10 |
| METTL4 | 18p11.32 | rs549315 | 2183055 | G/A | 0.378/0.429 | -0.29 | 0.0389 | -1.32 | 4.80E-10 |
| RGPD4 | 2q12.3 | rs700855 | 108368694 | T/C | 0.372/0.328 | 0.27 | 0.0454 | 1.31 | 9.91E-10 |
| RGPD4 | 2q12.3 | rs328025 | 108355045 | G/A | 0.377/0.324 | 0.31 | 0.0219 | 1.25 | 1.06E-09 |
| MALL | 2q13 | rs117802391 | 110862084 | C/T | 0.036/0.061 | -0.77 | 0.0198 | 19.57 | 1.06E-09 |
| LINC01978 | 17q25.3 | rs57851800 | 77896371 | A/C | 0.345/0.326 | 0.16 | 0.2657 | -1.27 | 1.08E-09 |
| RREB1 | 6p24.3 | rs9505086 | 7232186 | T/C | 0.286/0.307 | -0.11 | 0.4279 | 1.24 | 1.20E-09 |
| DST | 6p12.1 | rs117021265 | 56628021 | T/C | 0.034/0.024 | 0.25 | 0.5494 | -18.96 | 1.75E-09 |
| RPRM | 2q23.3 | rs5005908 | 154003680 | G/T | 0.344/0.27 | 0.32 | 0.022 | 1.26 | 3.03E-09 |
| FOXP1 | 3p13 | rs878118 | 71246228 | T/G | 0.238/0.255 | -0.15 | 0.323 | -1.39 | 3.85E-09 |
| LINC01344 | 1q25.3 | rs12033118 | 182229747 | C/T | 0.022/0.027 | -0.15 | 0.7192 | -19.75 | 5.00E-09 |
| RBMS3 | 3p24.1 | rs1979271 | 29607405 | T/A | 0.406/0.39 | 0.07 | 0.5841 | -1.17 | 6.47E-09 |
| CXCR4 | 2q22.1 | rs189432614 | 136809235 | A/G | 0.016/0.025 | -1.63 | 0.0131 | -34.81 | 7.59E-09 |
| CDH13 | 16q23.3 | rs3848296 | 82550548 | G/A | 0.192/0.231 | -0.16 | 0.3193 | 1.46 | 7.69E-09 |
| RUFY1 | 5q35.3 | rs4075890 | 178997373 | T/C | 0.216/0.2 | 0.1 | 0.5444 | 1.46 | 8.47E-09 |
| EIF2B5 | 3q27.1 | rs4350902 | 184352200 | T/C | 0.472/0.492 | -0.09 | 0.5253 | -1.18 | 8.83E-09 |
| PLEKHA1 | 10q26.13 | rs10510110 | 124192430 | C/T | 0.399/0.372 | 0.17 | 0.2098 | 1.26 | 8.96E-09 |
| PFKP | 10p15.2 | rs58183624 | 3107217 | C/T | 0.066/0.044 | 0.15 | 0.592 | -2.68 | 9.52E-09 |
| TRIM22 | 11p15.4 | rs7480654 | 5722839 | T/C | 0.317/0.284 | 0.18 | 0.22 | -1.21 | 9.59E-09 |
| LINC00879 | 3q11.2 | rs4411883 | 94549686 | T/G | 0.09/0.111 | -0.34 | 0.1404 | -1.91 | 1.24E-08 |
| TNIK | 3q26.31 | rs11925024 | 171014067 | A/C | 0.145/0.151 | -0.15 | 0.4431 | -0.9 | 0.0008 |
| TNIK | 3q26.31 | rs1231 | 171031233 | A/T | 0.144/0.154 | -0.18 | 0.3537 | -0.8 | 0.0021 |
| FTO | 16q12.2 | rs9302654 | 54009545 | C/T | 0.114/0.144 | -0.34 | 0.088 | 0.58 | 0.0334 |
| SLFN11 | 17q12 | rs77814639 | 33678827 | A/G | 0.184/0.153 | 0.28 | 0.1329 | -0.43 | 0.108 |
| SAP18 | 13q12.11 | rs9509543 | 21692404 | C/T | 0.346/0.356 | -0.05 | 0.705 | -0.2 | 0.2731 |
| EDNRA | 4q31.22 | rs6841581 | 148401190 | A/G | 0.13/0.217 | 0.53 | 0.0006 | -0.27 | 0.301 |
| SLC7A10 | 19q13.11 | rs11672303 | 33726375 | T/C | 0.171/0.154 | 0.08 | 0.6556 | 0.23 | 0.3238 |
| CACUL1 | 10q26.11 | rs11198727 | 120767097 | A/G | 0.382/0.429 | -0.13 | 0.3308 | 0.17 | 0.3558 |
| MPDZ | 9p23 | rs1332064 | 12942764 | T/C | 0.354/0.309 | 0.19 | 0.1724 | 0.1 | 0.5916 |
| UNC13C | 15q21.3 | rs4774715 | 55140204 | C/T | 0.432/0.441 | -0.01 | 0.9362 | -0.09 | 0.6185 |
| RYK | 3q22.2 | rs74585958 | 133773362 | G/A | 0.054/0.041 | 0.57 | 0.0817 | -0.21 | 0.6852 |
| EIF4H | 7q11.23 | rs150664966 | 73594157 | T/C | 0.016/0.022 | -0.17 | 0.7152 | -0.05 | 0.9384 |
* These were estimated by generalized linear model after adjusting for age, sex, hypertension, diabetes, hyperlipidemia, and smoking in the previous GWAS.
Table 2.
| Gene | Chr | SNP | BP | M/m | MAF, case/control | lnOR* | p-value for GWAS* | lnOR† | p-value for interaction† |
|---|---|---|---|---|---|---|---|---|---|
| EDNRA | 4q31.22 | rs6841581 | 148401190 | A/G | 0.13/0.217 | 0.53 | 0.0006 | NA | NA |
| RYK | 3q22.2 | rs74585958 | 133773362 | G/A | 0.054/0.041 | 0.57 | 0.0817 | -19.91 | 1.64E-09 |
| SAP18 | 13q12.11 | rs9509543 | 21692404 | C/T | 0.346/0.356 | -0.05 | 0.705 | 1.85 | 2.87E-09 |
| SLC7A10 | 19q13.11 | rs11672303 | 33726375 | T/C | 0.171/0.154 | 0.08 | 0.6556 | 2.16 | 3.55E-09 |
| FTO | 16q12.2 | rs9302654 | 54009545 | C/T | 0.114/0.144 | -0.34 | 0.088 | -3.1 | 3.78E-09 |
| SLFN11 | 17q12 | rs77814639 | 33678827 | A/G | 0.184/0.153 | 0.28 | 0.1329 | -18.5 | 4.48E-09 |
| EIF4H | 7q11.23 | rs150664966 | 73594157 | T/C | 0.016/0.022 | -0.17 | 0.7152 | 20.91 | 4.80E-09 |
| MPDZ | 9p23 | rs1332064 | 12942764 | T/C | 0.354/0.309 | 0.19 | 0.1724 | 1.75 | 5.10E-09 |
| UNC13C | 15q21.3 | rs4774715 | 55140204 | C/T | 0.432/0.441 | -0.01 | 0.9362 | -1.68 | 7.74E-09 |
| CACUL1 | 10q26.11 | rs11198727 | 120767097 | A/G | 0.382/0.429 | -0.13 | 0.3308 | 1.72 | 8.06E-09 |
| TNIK | 3q26.31 | rs11925024 | 171014067 | A/C | 0.145/0.151 | -0.15 | 0.4431 | -2.71 | 1.04E-08 |
| TNIK | 3q26.31 | rs1231 | 171031233 | A/T | 0.144/0.154 | -0.18 | 0.3537 | -2.86 | 1.22E-08 |
| METTL4 | 18p11.32 | rs549315 | 2183055 | G/A | 0.378/0.429 | -0.29 | 0.0389 | 0.59 | 0.033 |
| PTCH1 | 9q22.32 | rs1105980 | 98113635 | G/C | 0.27/0.294 | -0.08 | 0.5821 | -0.6 | 0.058 |
| MALL | 2q13 | rs117802391 | 110862084 | C/T | 0.036/0.061 | -0.77 | 0.0198 | -1.58 | 0.0762 |
| CXCR4 | 2q22.1 | rs189432614 | 136809235 | A/G | 0.016/0.025 | -1.63 | 0.0131 | 1.02 | 0.2116 |
| LINC01344 | 1q25.3 | rs12033118 | 182229747 | C/T | 0.022/0.027 | -0.15 | 0.7192 | 0.79 | 0.2679 |
| BOLL | 2q33.1 | rs700651 | 198631714 | A/G | 0.476/0.449 | 1.42 | 0.0079 | -0.27 | 0.301 |
| LINC01978 | 17q25.3 | rs57851800 | 77896371 | A/C | 0.345/0.326 | 0.16 | 0.2657 | -0.29 | 0.3135 |
| PLEKHA1 | 10q26.13 | rs10510110 | 124192430 | C/T | 0.399/0.372 | 0.17 | 0.2098 | -0.26 | 0.335 |
| PFKP | 10p15.2 | rs58183624 | 3107217 | C/T | 0.066/0.044 | 0.15 | 0.592 | 0.56 | 0.3378 |
| LINC00457 | 13q13.2 | rs1536847 | 35106975 | G/T | 0.295/0.329 | -0.18 | 0.2033 | 0.25 | 0.3692 |
| TRIM22 | 11p15.4 | rs7480654 | 5722839 | T/C | 0.317/0.284 | 0.18 | 0.22 | -0.26 | 0.3718 |
| CDH13 | 16q23.3 | rs3848296 | 82550548 | G/A | 0.192/0.231 | -0.16 | 0.3193 | -0.31 | 0.3982 |
| FOXP1 | 3p13 | rs878118 | 71246228 | T/G | 0.238/0.255 | -0.15 | 0.323 | 0.25 | 0.4035 |
| LINC00879 | 3q11.2 | rs4411883 | 94549686 | T/G | 0.09/0.111 | -0.34 | 0.1404 | -0.37 | 0.4182 |
| RGPD4 | 2q12.3 | rs328025 | 108355045 | G/A | 0.377/0.324 | 0.31 | 0.0219 | -0.19 | 0.4897 |
| RUFY1 | 5q35.3 | rs4075890 | 178997373 | T/C | 0.216/0.2 | 0.1 | 0.5444 | -0.21 | 0.5058 |
| RGPD4 | 2q12.3 | rs700855 | 108368694 | T/C | 0.372/0.328 | 0.27 | 0.0454 | -0.18 | 0.5209 |
| CCDC3 | 10p13 | rs12412014 | 12911725 | G/C | 0.281/0.291 | -0.09 | 0.5145 | -0.14 | 0.6273 |
| RPRM | 2q23.3 | rs5005908 | 154003680 | G/T | 0.344/0.27 | 0.32 | 0.022 | -0.13 | 0.6388 |
| RBMS3 | 3p24.1 | rs1979271 | 29607405 | T/A | 0.406/0.39 | 0.07 | 0.5841 | 0.13 | 0.6541 |
| RREB1 | 6p24.3 | rs9505086 | 7232186 | T/C | 0.286/0.307 | -0.11 | 0.4279 | 0.06 | 0.825 |
| DST | 6p12.1 | rs117021265 | 56628021 | T/C | 0.034/0.024 | 0.25 | 0.5494 | 0.16 | 0.8283 |
| C5orf60 | 5q35.3 | rs62405726 | 179069468 | G/A | 0.318/0.287 | 0.13 | 0.3646 | -0.02 | 0.9398 |
| EIF2B5 | 3q27.1 | rs4350902 | 184352200 | T/C | 0.472/0.492 | -0.09 | 0.5253 | -0.01 | 0.9793 |
* These were estimated by generalized linear model after adjusting for age, sex, hypertension, diabetes, hyperlipidemia, and smoking in the previous GWAS.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download