INTRODUCTION
MATERIALS AND METHODS
1. Animals
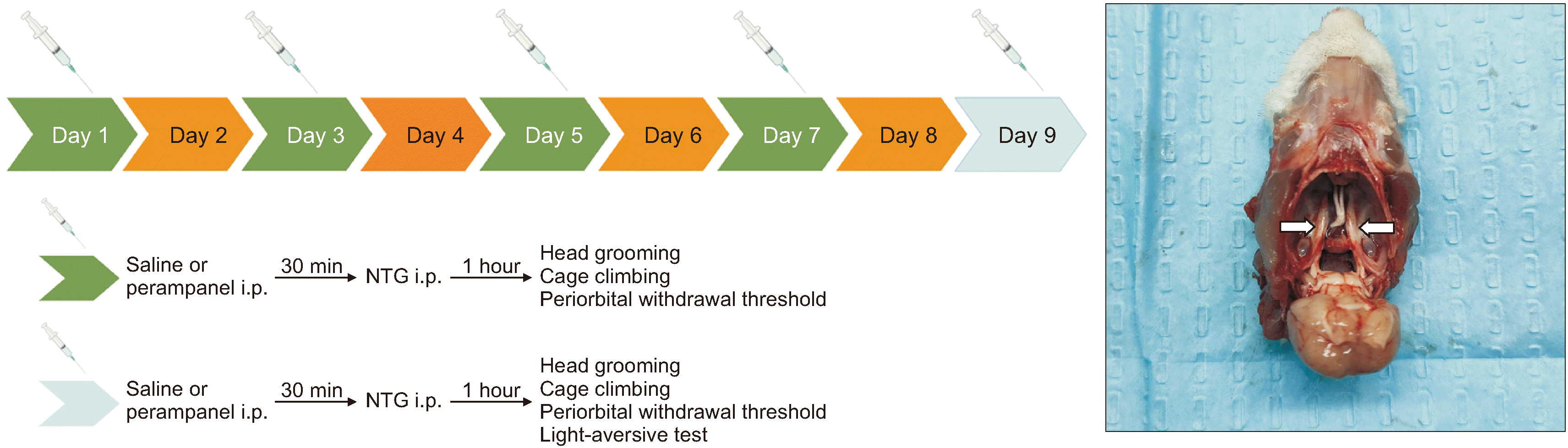
2. Experimental groups
3. Migraine induction and drug administration
4. Nociceptive behavior test
5. Light-aversive behavior test
6. Periorbital withdrawal threshold test
7. Body weight and food consumption measurement
8. Western blot
9. RT-qPCR
10. ELISA
11. HT-22 cells
12. Statistical analysis
RESULTS
1. Perampanel inhibited NTG-induced photophobia, migraine-like pain and nociceptive behaviors
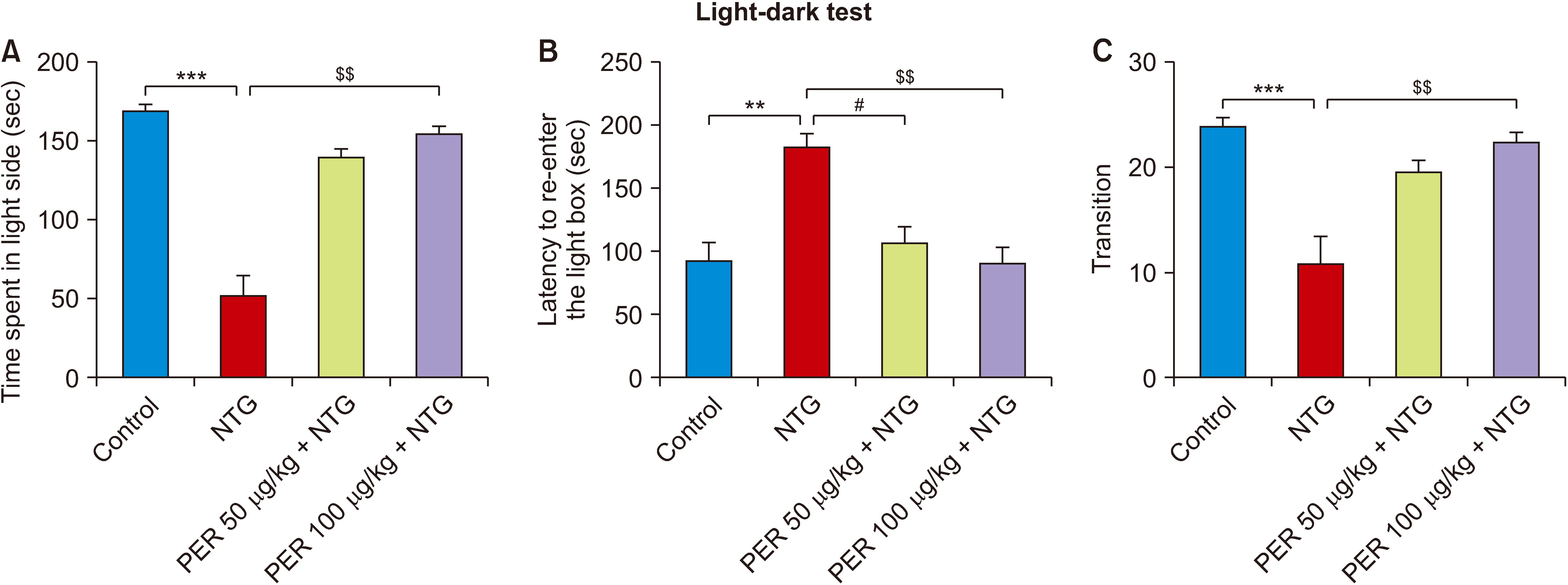 | Fig. 2Effects of perampanel treatment on photophobia. (A) Time spent in the light compartment, Kruskal–Wallis statistic = 24.30, ***P < 0.001; (B) Latency to re-enter the light compartment, Kruskal–Wallis statistic = 16.33, ***P = 0.001; (C) Transition, Kruskal–Wallis statistic = 20.55, ***P < 0.001. Control, n = 8; NTG, n = 8; PER 50 μg/kg + NTG, n = 8; PER 100 μg/kg + NTG, n = 8. Data are presented as means ± SEM. NTG: nitroglycerin, PER: perampanel. $$P < 0.01 vs. NTG group; **P < 0.01, ***P < 0.001 vs. control group; #P < 0.05 vs. NTG group in corresponding days. |
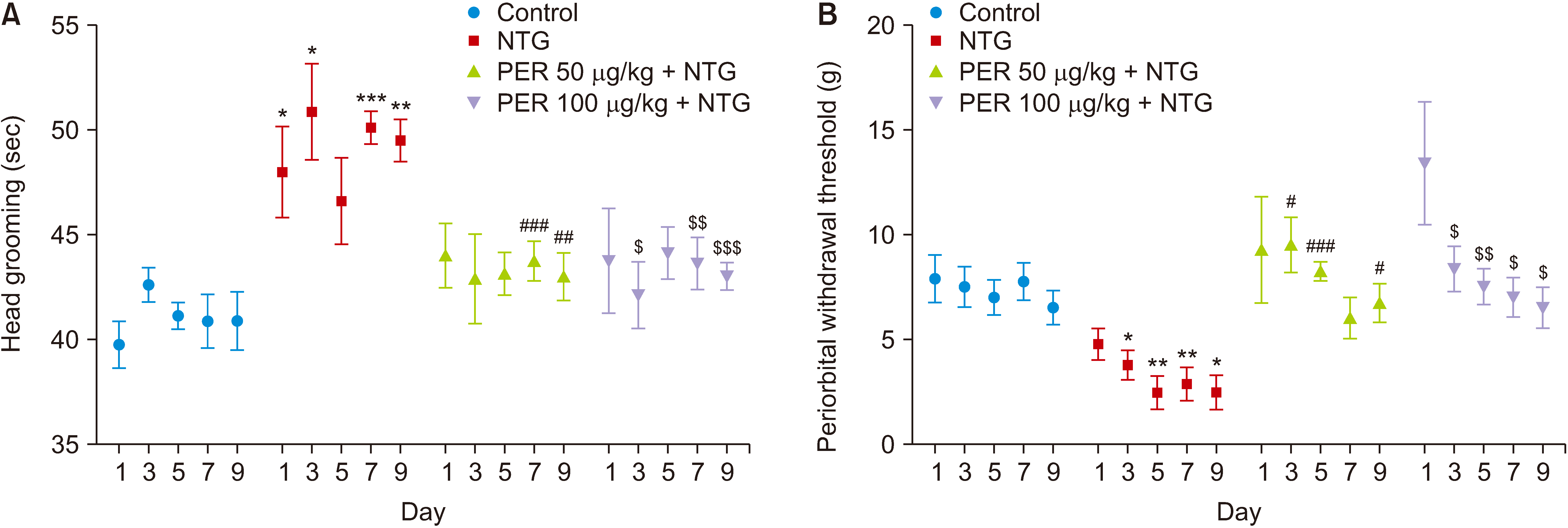 | Fig. 3Effects of perampanel treatment on NTG-induced migraine-like pain and nociceptive behaviors. (A) Head grooming, subgroups, F (3, 28) = 25.95, P < 0.001; time, F (4, 112) = 0.295, P = 0.880; subgroups × time, F (12, 112) = 0.629, P = 0.814. Control, n = 8; NTG, n = 8; PER 50 μg/kg + NTG, n = 8; PER 100 μg/kg + NTG, n = 8. In periorbital withdrawal thresholds test, NTG were decreased periorbital withdrawal thresholds, while perampanel treatment, at both doses of 50 and 100 μg/kg significantly increased the thresholds. (B) Periorbital withdrawal thresholds, subgroups, F (3, 28) = 23.28, P < 0.001; time, F (4, 112) = 4.397, P = 0.002; subgroups × time, F (12, 112) = 0.9606, P = 0.491. Data are presented as means ± SEM. NTG: nitroglycerin, PER: perampanel. $P < 0.05, $$P < 0.01, $$$P < 0.001 vs. NTG group; *P < 0.05, **P < 0.01, ***P < 0.001 vs. control group; #P < 0.05, ##P < 0.01, ###P < 0.001 vs. NTG group in corresponding days. |
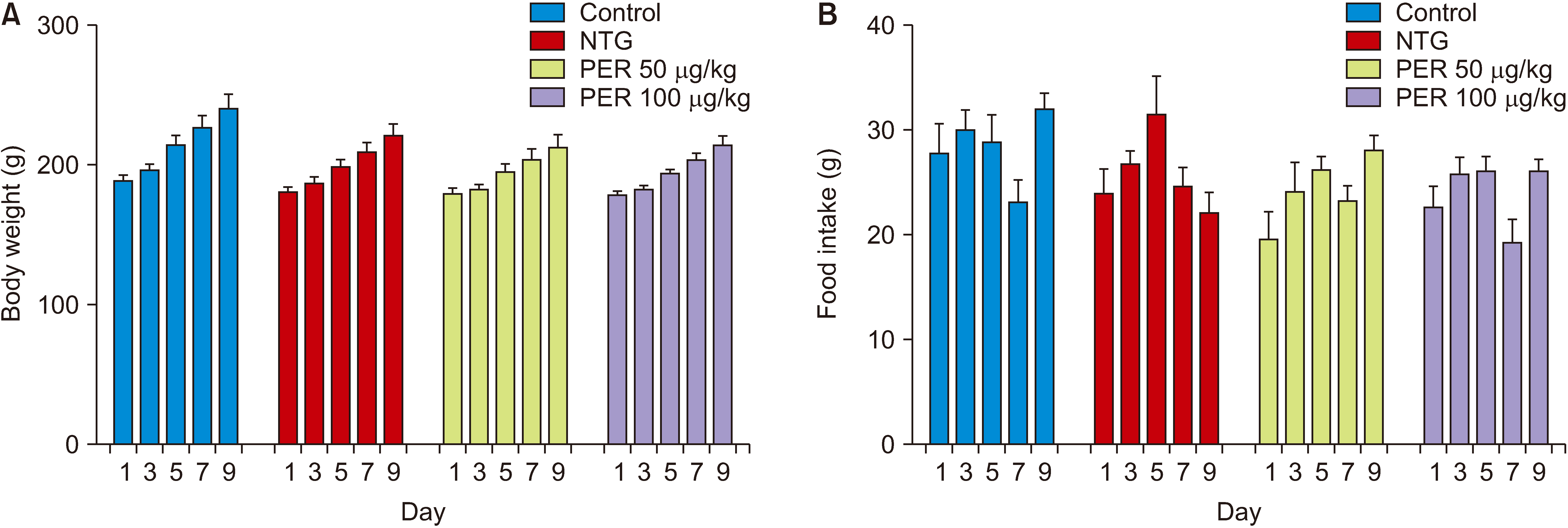 | Fig. 4Effects of perampanel treatment on eating disorders. (A) Body weight, subgroups, F (3, 28) = 4.720, P = 0.009; time, F (4, 112) = 67.16, P < 0.001; subgroups × time, F (12, 112) = 0.652, P = 0.793; (B) Food intake, subgroups, F (3, 28) = 4.311, P = 0.013; time, F (4, 112) = 5.916, P < 0.001; subgroups × time, F (12, 112) = 1.637, P = 0.091. Control, n = 8; NTG, n = 8; PER 50 μg/kg + NTG, n = 8; PER 100 μg/kg + NTG, n = 8. Data are presented as means ± SEM. NTG: nitroglycerin, PER: perampanel. |
2. Perampanel inhibited the increase in PACAP level
 | Fig. 5Effects of perampanel treatment on PACAP level in NTG-induced migraine. (A) Western blot showing the protein levels of PACAP; (B) Western bolt analysis of PACAP expression in TG. Kruskal–Wallis statistic = 12.15, **P = 0.007; (C) qRT-PCR analysis of PACAP gene expression in TG. Kruskal–Wallis statistic = 14.16, **P = 0.003, P < 0.001; (D) Elisa analysis of PACAP level in serum. Kruskal–Wallis statistic = 9.828, *P = 0.020. Control, n = 4; NTG, n = 6; PER 50 μg/kg + NTG, n = 6; PER 100 μg/kg + NTG, n = 6. Data are presented as mean ± SEM. PACAP: pituitary adenylate-cyclase-activating polypeptide, NTG: nitroglycerin, TG: trigeminal ganglion, PER: perampanel. Significant differences: *P < 0.05, ***P < 0.001. |
3. Perampanel affected the PKA/CREB signaling pathways in NTG-induced migraine
 | Fig. 6Effects of perampanel on PKA/CREB and PLC/PKC signaling pathways in NTG-induced migraine. (A) Western blot showing the protein levels of PKA, CREB and p-CREB; (B) Western blot analysis of the expression of PKA in TG. Kruskal–Wallis statistic = 17.20, ***P < 0.001; (C) Western blot analysis of the expression of P-CREB in TG. Kruskal–Wallis statistic = 11.52, **P = 0.009; (D) Western blot analysis of the expression of CREB in TG. Kruskal–Wallis statistic = 9.125, *P = 0.028; (E) Western blot showing the protein levels of PLC and PKC. (F) Western blot analysis of the expression of PLC in TG. Kruskal–Wallis statistic = 3.631, P = 0.304; (G) Western blot analysis of the expression of PKC in TG. Kruskal–Wallis statistic = 7.030, P = 0.071. Control, n = 4; NTG, n = 6; PER 50 μg/kg + NTG, n = 6; PER 100 μg/kg + NTG, n = 6. Data are presented as mean ± SEM. PKA: protein kinase A, CREB: cAMP-responsive-element-binding protein, PLC: phospholipase C, PKC: protein kinase C, NTG: nitroglycerin, TG: trigeminal ganglion, PER: perampanel. Significant differences: *P < 0.05, **P < 0.01. |
4. Perampanel decreased the expression of PACAP via inhibition of the cAMP/PKA/CREB pathway in vitro
 | Fig. 7Effects of perampanel on cAMP/PKA/CREB signaling pathways in vitro. (A) Western blot showing the protein levels of PACAP in HT-22 cell incubated with forskolin 10 µM for 24 hours; (B) Western blot analysis of the expression of PACAP. Kruskal–Wallis statistic = 7.538, *P = 0.011. Control, n = 4; Forskolin 10 µM, n = 4; PER 100 μM, n = 4; (C) Western blot showing the protein levels of PACAP in HT-22 cell incubated with 8-bromo-cAMP sodium salt 0.1 mM for 24 hours; (D) Western blot analysis of the expression of PACAP. Kruskal–Wallis statistic = 16.33, P = 0.001. Control, n = 4; 8-bromo-cAMP sodium salt 0.1 mM, n = 4; PER 100 μM, n = 4; (E) Western blot showing the protein levels of PACAP in HT-22 cell incubated with SQ22536 100 μM for 24 hours; (F) Western blot analysis of the expression of PACAP. Kruskal–Wallis statistic = 9.269, ***P < 0.001. Control, n = 4; SQ22536 100 μM, n = 4; PER 100 μM, n = 4. (G) Western blot showing the protein levels of PACAP in HT-22 cell incubated with H-89 10 µM for 24 hours; (H) Western blot analysis of the expression of PACAP. Kruskal–Wallis statistic = 9.846, ***P < 0.001. Control, n = 4; H-89 10 µM, n = 4; PER 100 μM, n = 4. (I) Western blot showing the protein levels of PACAP in HT-22 cell incubated with KG501 25 µM for 24 hours; (J) Western blot analysis of the expression of PACAP. Kruskal–Wallis statistic = 8.769, **P = 0.001. Control, n = 4; KG501 25 µM, n = 4; PER 100 μM, n = 4. Data are presented as mean ± SEM. PKA: protein kinase A, CREB: cAMP-responsive-element-binding protein, PER: perampanel. Significant differences: *P < 0.05, **P < 0.01. |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download