Abstract
Ion homeostasis, which is regulated by ion channels, is crucial for intracellular signaling. These channels are involved in diverse signaling pathways, including cell proliferation, migration, and intracellular calcium dynamics. Consequently, ion channel dysfunction can lead to various diseases. In addition, these channels are present in the plasma membrane and intracellular organelles. However, our understanding of the function of intracellular organellar ion channels is limited. Recent advancements in electrophysiological techniques have enabled us to record ion channels within intracellular organelles and thus learn more about their functions. Autophagy is a vital process of intracellular protein degradation that facilitates the breakdown of aged, unnecessary, and harmful proteins into their amino acid residues. Lysosomes, which were previously considered protein-degrading garbage boxes, are now recognized as crucial intracellular sensors that play significant roles in normal signaling and disease pathogenesis. Lysosomes participate in various processes, including digestion, recycling, exocytosis, calcium signaling, nutrient sensing, and wound repair, highlighting the importance of ion channels in these signaling pathways. This review focuses on different lysosomal ion channels, including those associated with diseases, and provides insights into their cellular functions. By summarizing the existing knowledge and literature, this review emphasizes the need for further research in this field. Ultimately, this study aims to provide novel perspectives on the regulation of lysosomal ion channels and the significance of ion-associated signaling in intracellular functions to develop innovative therapeutic targets for rare and lysosomal storage diseases.
Lysosomes play vital roles in maintaining extracellular and intracellular macromolecules while regulating various degradation processes. They modulate ion-related functions within endocytic, exocytic, and other signaling pathways, thereby contributing to cellular signaling. Lysosomes are distributed throughout the body, and defects in lysosomal function are associated with lysosomal storage disorders (LSDs). To regulate lysosomal function, ion channels, and transporters are necessary to maintain ion concentration gradients of Na+, K+, Ca2+, H+, and Cl-. Each ion channel operates independently and in correlation with the others, responding to changes in individual ion concentrations and aiding in the establishment of the lysosomal membrane potential. Lysosomes maintain an acidic pH range (approximately 4.5–5.5) to support the enzymatic activity required for degradative functions. Ion channel activity influences acidic conditions and plays crucial roles in diverse cellular processes, including exocytosis, calcium release, wound repair, and recycling. Autophagy and cellular signaling can be severely affected by lysosome dysfunction. Dysfunctions in lysosomal ion channels contribute to the development of various diseases, including LSDs, neurodegenerative diseases, metabolic diseases, such as Mucolipidosis IV (MLIV), and autoimmune diseases. This highlights the importance of identifying novel therapeutic targets as increased lysosomal dysfunction may contribute to the prevalence of these diseases.
Numerous reviews have indicated the potential of lysosomal ion channels as therapeutic targets in these diseases [1-3]. In the context of the Coronavirus disease 2019 (COVID-19) era, the lysosomal two-pore channel (TPC) has emerged as a novel target for regulating virus-associated diseases such as severe acute respiratory syndrome (SARS), Ebola, and COVID-19.
The first descriptions of LSD phenotypes were made in the 19th century [4,5]. However, it wasn't until 1955–1956 that lysosomes were identified. de Duve [6] played a crucial role in this discovery by identifying lysosomes as intracellular catabolic stations. This has marked the beginning of extensive research on lysosomes. In one of his studies, he examined the localization of glucose-6 phosphatase in hepatic fractions, thereby providing mechanistic insights. This allowed him to isolate and identify non-specific acid phosphatases and glucose-6-phosphate. Christian discovered acid phosphatase-rich regions between microsomes and mitochondrial fractions, which he termed the "L-fraction" [7]. They named it the lysosome and hypothesized it functioned as a protein degradative organelle. Initially, lysosomes were considered garbage boxes that were primarily involved in recycling and digestion. However, recent studies have provided new insight into their roles in autophagy and mitochondrial function. Furthermore, there has been a renewed emphasis on understanding their function as nutrient sensors.
Numerous lysosomal-resident proteins play significant roles in cellular signaling pathways associated with organelle function. Approximately 50 proteins were categorized as acidic hydrolases [8-10]. Most of these proteins are localized within the lysosomal lumen and function as sulphatases or exoglycosidases. However, some proteins found in the lysosomal membrane contribute to various functions, including acidification, transport, facilitation of interactions between lysosomes and other organelles, and maintenance of membrane stability [11,12]. Notably, lysosomal proteins do not operate in isolation but rather interact with other proteins. These interactions are essential for preserving the integrity of lysosomes and ensuring the proper functioning of lysosomes, both internally and externally. Some of the proteins involved in these interactions include protein kinase B (AKT), mechanistic targets of rapamycin complex 1 and 2 (mTORC1 and mTORC2), transcription factor EB (TFEB), folliculin (FLCN), FLCN-interacting protein, signal transducer and activator of transcription-3, and energy-sensing complex AMP-activated kinase (AMPK) [13-29].
Lysosomes play a primary role in the degradation process by influencing various components such as nucleic acids, sphingolipids, proteins, glycosaminoglycans, and complex lipids. They break down the extracellular and intracellular materials that are old or no longer useful, resulting in the production of amino acids, fatty acids, and saccharides. These dissolved components and substrates are then directed to their designated destinations via diverse routes, such as endocytosis, exocytosis, phagocytosis, and autophagy [2,30-32].
Autophagy, also known as the autophagy-lysosomal pathway, plays a crucial role in regulating cellular processes, such as intracellular clearance, recycling of molecules and cellular components, and preservation of intracellular energy metabolism. It involves the formation of phagophores and the recruitment of cargo, eventually leading to the fusion of lysosomes to form autolysosomes [9,16]. Lysosomal degradation is essential for autophagy and the elimination of organelles from cells. Defective lysosomes can induce cytotoxicity and trigger unexpected inflammatory responses. Over 60 known nucleases, phosphatases, lipases, and proteases degrade organelles and proteins through degradation steps [33-35]. During these degradation steps, lysosomes release essential components synthesized from new molecules or associated with cellular signaling pathways via the diffusion or transport of proteins, such as the cholesterol transporters Niemann-Pick type C protein 1, 2 (NPC1, 2) and nicotinic acid adenine dinucleotide phosphate (NAADP) [36-38].
An acidic environment within lysosomes is critical for activating lysosomal enzymes and facilitating the conversion of their pro-forms into the mature forms necessary for degradation. Lysosomal enzymes involved in the degradation process are highly pH-dependent and are influenced by designated pH regulation for proper functioning. Disruptions in pH can interfere with substrate clearance, potentially linking diseases such as Parkinson's disease (PD) and MLIV, in which abnormal autophagy pathways contribute to lysosomal damage and improper protein aggregation.
Recently, lysosomes have been recognized as nutrient sensors that monitor cellular nutrient conditions and help maintain stable intracellular metabolism by adjusting various components.
Lysosomal ion channels have only been discovered in the past decade, posing challenges in their study owing to the limitations of electrophysiological techniques. However, these channels play crucial roles in maintaining lysosomal homeostasis. By regulating ion channel activity, lysosomal dysfunction can be rescued, and promising therapeutic targets for diseases associated with impaired lysosomal function can be identified. Various techniques have been developed to investigate ion channel activity under different lysosomal conditions. Specific ions, such as Na+, K+, Ca2+, and Cl-, have been identified as key players in the maintenance of lysosomal homeostasis. Although the number of reported lysosomal ion channels is limited, their dysfunction has been strongly linked to impaired lysosomal function. This review focuses on the functions of lysosomal TPC, calcium-activated large-conductance K+ channel (BK), TMEM175, ClC-7, and TRPML channels (Fig. 1) and highlights their potential implications in lysosomal-related diseases.
TPCs in lysosomal function: TPCs are characterized by a dimeric structure with two-pore domains. Three types of TPC channels have been identified: TPC1, TPC2, and TPC3 (absent in mice, rats, and primates). TPC3 is currently under investigation because it localizes to the plasma membrane and is found in Na+-selective and Ca2+-permeable endosomal and lysosomal membranes [39-41]. TPC1, in contrast, is a voltage-dependent channel that is activated by PI(3,5)P2, the second messenger of NAADP, and changes in pH [41-45]. TPC2, although independently activated by voltage [40,42,46], has caused some problems regarding its activator. Some studies have suggested that TPC channels are activated by PI(3,5)P2 and increase Na+ influx [43], whereas others have proposed that NAADP activates Ca2+ influx, which is associated with TFEB translocation [47]. Structural studies conducted by Guo et al. [47] have indicated that TPC channels exhibit Na+ and Ca2+ sensitivity. PI(3,5)P2 has been shown to directly bind to TPCs and open channels, based on structural findings [45,48,49]. However, NAADP activation remains a subject of debate and requires further investigation, as it possibly differs among various organs. TPCs function as intracellular sensors during nutrient deprivation [43] and interact directly with mTOR, influencing channel activation and lysosomal function based on nutrient status. They are involved in pH homeostasis and membrane trafficking, affecting the pathogenicity of viruses, such as pigmentation-related diseases, Ebola, SARS, and COVID-19 [50-53]. TPC1 also plays a crucial role in the endosomes and participates in protein processing and uptake. Defects in TPC1 hinder toxin and protein uptake by weakening their interaction with syntaxins (SNARE syntaxins 7 and 8), leading to aberrant autophagy pathways due to lysosomal dysfunction from endosomes to lysosomes. TPC2 regulated lysosomal pH and activity. Overexpression of TPC2 led to an increase in lysosomal pH, suggesting its involvement in lysosomal homeostasis. Additionally, it modulates lysosomal membrane potential and channel activity [43,54]. Some studies have demonstrated that TPC2 knockout fibroblasts do not exhibit changes in lysosomal pH, but reveal increased lysosomal Ca2+ levels and reduced cholesterol degradation during low-density lipoprotein (LDL)-derived cholesterol transport [55,56]. Disruption of TPC activity can affect endosomal trafficking, thereby inhibiting viral spread [57,58], indicating a role for TPCs in lysosomal trafficking. Collectively, the regulation of lysosomal ion channels represents a promising novel therapeutic target.
Potassium influx helps maintain homeostasis by compensating for the loss of Na+ and Ca2+. Similar to the plasma membrane, the movement of K+ ions is closely linked to lysosomal membrane potential to ensure normal lysosomal function. Although the BK is believed to be the sole K+ channel in endolysosomes, a novel transmembrane protein, TMEM175, has been identified as a potassium-selective channel [13].
TMEM175: TMEM175 localizes within endolysosomes and was initially believed to exist only in prokaryotes. However, after the development of mass spectrometry, it was recognized as a mammalian protein [59]. TMEM175 acts as a K+ channel with the selective movement of K+ ions [60]. Unlike canonical K+ channels, TMEM175 does not inhibit tetraethylammonium (TEA) or quinine but suppresses Zn2+ and 4-AP. Although a P-loop selectivity filter is lacking in commercial K+ channels, structural studies have suggested that threonine and serine residues play a role in K+ selectivity [61]. Defects in TMEM175 have been found to regulate lysosomal pH and autophagy stages in RAW246.7 macrophage cells [60]. TMEM175 is strongly associated with neurodegenerative diseases like PD and may be involved in various LSDs [62,63]. It has emerged as a major protein connecting these diseases, as revealed by studies comparing Lewy body dementia patients and controls [62,64]. Additionally, TMEM175 deficiency affected mitochondrial function and autophagy in neuroblastoma cells [65] (Fig. 2).
BKs: In contrast, BK channels are calcium-activated large-conductance channels comprising six transmembrane segments and four pore-forming BK α-subunits (BKβ1-4). These channels were initially identified at the Slowpoke locus Slo1 in Drosophila [66]. BK channels not only respond to cytosolic Ca2+ but also influence membrane depolarization. Unlike other channels, BK channels are widely expressed and have been extensively studied in the plasma membrane. When activated, they facilitate K+ export from the cytosol, leading to hyperpolarization and increased lysosomal membrane. Some studies have suggested that BK channels are associated with TRPML1-mediated Ca2+ release. TRPML1 activation leads to Ca2+ release, which subsequently activates BK channels, allowing the influx of K+ ions into the lysosomal lumen [67]. This sustains the membrane potential necessary for the continuous activation of TRPML1 and V-ATPase. Inhibition of BK channels reduces lysosomal Ca2+ release, but co-treatment with a TRPML1 activator can rescue this effect.
TRPML channels: function, localization, and implications in lysosomal disorders: TRPML channels, initially discovered in Drosophila melanogaster, belong to the transient receptor potential (TRP) superfamily. These channels are nonselective cation channels that allow the permeation of calcium ions. The TRP superfamily comprises six main subgroups based on their functions: TRPA (ankyrin), TRPC (canonical), TRPM (melastatin), TRPML (mucolipin), TRPP (polycystin), and TRPV (vanilloid) [68,69]. TRPML isoforms include TRPML1-3, which are six transmembrane proteins with N- and C-termini. TRPML1 and TRPML2 are located on chromosome 19, whereas TRPML3 is located on chromosome 1 [70-72]. These isoforms share approximately 40% amino acid sequence similarity [73,74], and the localization of TRPML3 depends on the presence of TRPML1 and TRPML2. Controversially, it has been reported that both TRPML1 and TRPML2 bind to TRPML3 [75].
Among TRPML channels, TRPML1 is widely expressed [55], and localized in endosomes and lysosomes. The activation of TRPML1 channels leads to increased ion transport from the lysosomal lumen to the cytosol [76]. These channels are activated by PI(3,5)P2 and ML-SA1 but inhibited by ML-SL1. TRPML channels function within the lysosomes and interact with each other. TRPML1 has been extensively studied because of its association with LSDs. Mutations in TRPML1, also known as MCOLN1-3, are linked to LAMP-1 in the lysosomal membrane [77,78]. TRPML1 defects disrupt lysosomal conditions and lead to autophagic dysfunction and neurodegeneration. This highlights the importance of TRPML1 in organelle homeostasis and cellular function and suggests its role in lysosomal pH regulation. Interestingly, TRPML1 exhibited the opposite behavior of other ion channels. Dysfunction of TPCs and TMEM175 or nutrient deficiency increases lysosomal pH [13,42,79], whereas TRPML1 moves towards an acidic pH when depleted. TRPML1 plays a crucial role in sustaining the lysosomal function by interacting with lysosomal enzymes. However, some studies have suggested that pH can also influence TRPML channels [74]. TRPML1 contains an intraluminal pore with aspartate residues that modulate Ca2+ conductance depending on the surrounding pH [79].
TRPML2 and TRPML3: tissue localization and functions: TRPML2 exhibits tissue-specific expression and is primarily found in the heart, kidney, thymus, liver, immune cells, endosomes, and lysosomes [80]. Although the exact functions of TRPML2 channels are not well understood, some studies have suggested their association with glycosylphosphatidylinositol-anchored proteins (GPI-APs), a major histocompatibility protein class I, and ADP-ribosylation factor (ARF6), a small G protein involved in membrane trafficking and recycling. TRPML2 localizes to the ARF6-modulated pathway and regulates GPI-AP sorting [81].
In contrast, TRPML3 is found in various tissues, including the spleen, lungs, eyes, thymus, and melanocytes [82,83]. It is primarily localized in the endosomes, lysosomes, and intracellular vesicles [84,85]. TRPML3 plays a crucial role in regulating sodium ions (Na+) and cytosolic hydrogen ions (H+). However, it exhibited distinct pH regulation compared to TRPML1. The functional pH of TRPML3 is approximately 6.4, and its activity is inhibited at an acidic pH [85]. TRPML3 is present in multiple cellular compartments including the plasma membrane, endocytosis, and autophagy. Its activity is inhibited by acidic pH; however, when overexpressed or activated, it enhances autophagy and regulates its status. TRPML3 interacted with GATE16, an ATG8 homolog [86]. During autophagy induction, TRPML3 undergoes palmitoylation, a post-translational modification that facilitates the movement of specific components within the autophagosome, activating the process [87]. Previous studies have suggested a correlation between the number of autophagosomes and TRPML3 expression during cellular stress and nutrient deprivation. Cells lacking TRPML3 show reduced autophagosome formation, potentially leading to nutrient deficiency [88]. TRPML3 also interacts with autophagosome markers, indicating its direct involvement in the regulation of Ca2+-mediated membrane fusion. Structural studies have identified specific PIP2 binding site mutations (K52AR58AK62A, R305A, and F524A) that abolish TRPML3 currents, whereas K52AR58AK62A and R305A completely block PIP2 binding [83]. Constitutively activating isoforms of TRPML3, including the A419P and I362T mutants, have been reported, although the status of I362T remains controversial. Variants such as A419P are associated with conditions such as deafness and vascular problems because these mutations cause continuous Ca2+ release from lysosomes into the cytosol, leading to elevated cytosolic Ca2+ levels and disruption of intracellular signaling pathways. In the A419P mutant, sensory hair cell death results in hearing loss. Another mutation, I362T, reduced TRPML3 currents, resulting in a milder phenotype when co-mutated with A419P. Recent findings indicated that phosphatidylinositol-3-phosphate (PI3P), a crucial lipid for autophagosome formation, specifically interacts with TRPML3. The binding of TRPML3 to PI3P increases Ca2+ release from phagophores and promotes autophagy. Furthermore, TRPML3 inhibition abolishes autophagy, even in the presence of PI3P [89].
The ClC family encompasses the voltage-gated Cl- channels that distinguish themselves from others by functioning as Cl-/H+ antiporters [90,91]. These channels are present in the plasma membrane and intracellular organelles. Depending on their location, ClC channels contribute to various processes, such as resting membrane potential, pH regulation, and overall homeostasis [90,91].
Among them, ClC-7 is widely expressed and prominently found in the central and peripheral nervous systems. It localizes to late endosomes and lysosomes, alongside Lamp-1 [92-99]. For its distinct functioning and localization, ClC-7 requires co-localization with the β subunit of osteoclastogenesis-associated transmembrane protein 1 [100,101]. ClC-7 is expressed at the ruffled borders of osteoclasts [92]. However, its specific role in the lysosomes remains debatable.
Recent findings have suggested that ClC-7 defects have a minimal impact on phagosomal acidification. Instead, ClC-7 primarily contributes to the accumulation of lysosomal Cl-, while V-ATPases are responsible for maintaining a low pH necessary for degradative hydrolase activity. ClC-7 indirectly activates V-ATPase by providing a driving force for Cl- accumulation [102].
Other studies have proposed that ClC-7 is predominantly associated with lysosomal hyperacidification, leading to the depletion of PI(3,5)P2 by inhibiting PIKfyve (an FYVE finger-containing phosphoinositide kinase) [65,103]. Notably, PIKfyve potently activates ClC-2 [104]. This suggests that ClC-7 is directly inhibited by PI(3,5)P2, thereby preventing functional mutations (such as Y715C) and regulating lysosomal pH [103,105].
LSDs are inherited metabolic disorders with a prevalence of approximately 1 in 5,000 live births [106], indicating the critical role of lysosomes in numerous cellular processes. Lysosomal dysfunction leads to various defects in lysosomal properties including pH imbalance, membrane potential disruption, enzymatic activity impairment, and abnormal autophagy. These lysosomal-mediated diseases encompass a broad range of conditions, from LSDs to neurodegenerative diseases, such as Parkinson's and Alzheimer’s disease, hearing loss, obesity, osteopetrosis, fatty liver, cancers (lung, breast, prostate, and colon), and viral diseases (Ebola and SARS) [107-118].
LSDs, which involve mutations or defective channel function, are common lysosomal dysfunctions. One such example is MLIV, a neurodegenerative and neurodevelopmental disease caused by mutations in TRPML1. Patients with MLIV exhibit dysfunctions in mTOR, trafficking, metabolism, lysosomes, and autophagy. Vardi et al. [119] conducted a study using TRPML1 knockout mice and obtained proteomic data through pathway analysis. The study revealed upregulated pathways in the cerebellum and cerebral cortex (integrin pathway, innate immune system, lysosome, and sphingolipid metabolism, among others) as well as downregulated pathways (GABAergic synapses). TRPML1 knockout mice displayed upregulated immune response, cytokine production, coagulation cascades, and viral susceptibility [119].
Niemann-Pick disease, another type of LSD, is associated with TRPML channels. This group of disorders causes cellular changes due to the accumulation of cholesterol and other lipids in endocytic compartments. In Niemann-Pick disease, cholesterol trafficking from the lysosome to the endoplasmic reticulum leads to dysfunction in transcriptional cholesterol metabolism and esterification. The improper regulation of lysosomal calcium release by TRPML1 mutations is thought to be associated with these issues [120].
Over the past few years, COVID-19 has become a pandemic, primarily causing respiratory syndromes related to SARS-CoV-2. While some individuals recover or improve naturally, others develop acute respiratory distress syndrome [121-123]. However, a few individuals may experience primary cardiovascular dysfunctions [124,125]. TPC channels, which localize in endosomes, lysosomes, and Na+ and Ca2+ channels, have been identified as potential candidates. It is suggested that TPCs regulate Ca2+ associated with SARS-CoV-2 through endocytosis and endolysosomal trafficking [50,126]. However, this hypothesis warrants further investigation. TPC1 is associated with systemic anaphylaxis and mast cell activity. TPC1-deficient mice exhibited enhanced passive systemic anaphylaxis, lower body temperature, slower recovery, reduced histamine release, decreased mast cell number and size, and impaired regulation of exocytosis [127]. TPC2, in contrast, modulates the endolysosomal degradation pathway by regulating trafficking. TPC2 dysfunction impairs epithelial growth factor (EGF)/EGF-receptor trafficking and LDL-derived cholesterol in mouse fibroblasts and hepatocytes. This indicates that TPC2 plays a role in regulating the trafficking mechanism of endolysosomal degradation pathways, which affect metabolite and macromolecule homeostasis. Recently, TPC2 has been shown to rescue lysosomal storage in MLIV and Batten disease. This channel activation improves the cellular phenotypes associated with lipofuscin accumulation, lactosylceramide buildup, cholesterol storage, and abnormal vacuole formation. TPC2 activation promotes lysosomal exocytosis and autophagy, providing a potential therapeutic avenue for these diseases [128].
Although BK channels were initially studied in the context of cancer, they have now gained attention for their use in neurological diseases. Mutations in KCNMA1, which encodes the pore-forming-subunit of BK channels, have been identified as crucial points. BK channel conductance plays a role in the regulation of breast cancer progression, and its localization and activation are linked to cancer development. Low expression of KCNMA1 has been associated with shorter recurrence-free survival in patients with breast tumors, regardless of treatment or estrogen receptor status. LRRC26, an auxiliary subunit, is required to activate BK channels in non-excitable tumor cells at a depolarized resting membrane potential [129]. In terms of neurological diseases, mutations in KCNMA1 (BKG354S) located in the selectivity filter and pore region have been linked to accelerated cognitive impairment in children with congenital and progressive cerebellar ataxia. These mutations significantly reduce ion selectivity and channel conductance and impair cell viability, mitochondrial content, and neurite outgrowth; however, they do not affect channel trafficking. However, the BK activator, NK1619, selectively blocks mutated BKG354S channels, suggesting a potential therapeutic strategy [130]. Another study proposed a correlation between KCNMA1 mutations and paroxysmal non-kinesigenic dyskinesia 3, which may occur with or without generalized epilepsy in mice and in three KCNMA1 patient variants. The BKN999S and BKD434G channels exhibited a gain-of-function phenotype, whereas the BKH444Q channel was negatively activated in heterologous cells. Heterozygous mutant mice with activating mutations show increased action potential firing and reduced seizure thresholds. However, this effect was not observed in mice harboring the H444Q mutation. N999S heterozygous mice demonstrate severe behavioral impairment in paroxysmal dyskinesia tests, suggesting its role in promoting seizure propensity (Table 1) [131].
TMEM175 has recently been implicated in PD. Unlike most ion channels that exhibit heterozygous activity, TMEM175 activation is dose-dependent. TMEM175 knockout mice display impaired clearance of phosphorylated α-synuclein (S129), a marker of PD, and increased galectin-3 levels, indicating lysosomal membrane damage. TMEM175 also functions as a nutrient sensor and its knockout negatively affects lysosomal pH, enzyme function, and autophagy. Furthermore, TMEM175 knockout mice were more susceptible to damage induced by starvation and neurotoxins (MPP+ and H2O2) than wild-type mice. Mutations in TMEM175 (M393T and Q65P) are associated with PD. The M393T mutation increased the likelihood of developing PD by more than 25%. Patients with PD and the M393T mutation experience decreased motor and cognitive functions. In vitro studies have shown that the M393T mutation reduces channel activity, impairs α-synuclein clearance, causes nutrient deficiency, and increases sensitivity to neurotoxins. In contrast, the Q65P mutation affected channel activity differently and was more resistant to nutrient deficiency. After deficiency, growth factors can rescue channel activity, indicating a connection between growth factors and TMEM175 channels. Protein kinase B (AKT), which is involved in multiple signaling pathways including cell proliferation, apoptosis, and metabolism, directly influenced TMEM175 activity through conformational changes in the AKT PH domain [13].
The industrial application of TMEM175 as a therapeutic target is supported by collaborations such as that between Caraway and AbbVie, which aim to develop therapies targeting TMEM175 with a funding allocation of up to $267 million. This highlights the potential significance of lysosomal ion channels as targets for therapeutic interventions. Another channel, TRPML1, has also been investigated as a potential target for agonist development to treat neurodegenerative diseases and muscular dystrophy. Merck has invested up to $576 million in this study owing to the close relationship between TRPML1 and lysosomal calcium. Reactive oxidative stress (ROS), TFEB, and GSK3 signaling are additional possible connections that warrant further investigation [132-138]. Reports have suggested that Mucopolysaccharidosis and Krabbe disease may be caused by ROS, indicating the potential involvement of TRPML and TPC channels. These channels can modulate lysosomal calcium release, affect local calcium levels, and potentially influence intracellular signaling. TFEB-mediated autophagy is linked to LSDs through the inhibition of mTOR pathways by drugs such as rapamycin. However, it has been observed that rapamycin can independently activate TRPML1 channels, leading to lysosomal calcium release and affecting TFEB. This interaction elucidates the coordination between lysosomal function and autophagy regulation. GSK3, a member of the AKT signaling pathway, is associated with familial Alzheimer's disease. Inhibition of GSK3 increases lysosomal activity, although its connection lies with plasma membrane ion channels rather than lysosomal ion channels.
The emerging understanding of lysosomal ion channels and their involvement in various diseases, such as LSDs and neurodegenerative conditions, opens exciting possibilities for the development of novel therapeutic strategies. Targeting lysosomal ion channels may provide a new approach to modulating lysosomal function, restoring cellular homeostasis, and alleviating disease symptoms.
Further research is required to elucidate the precise mechanisms and interactions of lysosomal ion channels with other proteins and signaling pathways. Investigating these complex networks can provide deep insights into underlying disease processes and identify novel therapeutic targets. Additionally, exploring the functional roles of lysosomal ion channels in different cellular contexts and organ systems can help to develop individualized and effective treatment approaches. Continuous research in this field holds significant potential for the development of innovative therapies that mitigate the impact of lysosomal dysfunction and neurodegeneration, thereby improving the quality of life of individuals suffering from these diseases.
Lysosomes play a crucial role in cellular signaling and maintaining cellular homeostasis. Dysfunctional lysosomes disrupt cellular homeostasis and contribute to the development of various diseases including neurodegenerative disorders and LSDs. Lysosomal ion channels, such as lysosomal membrane potential channels, are primarily responsible for regulating lysosomal conditions. The pathogenesis of these diseases has been linked to dysfunctions in these ion channels and their related signaling pathways. Therefore, a comprehensive understanding of the correlation between lysosomal ion channels, their regulatory mechanisms, and their impact on intracellular signaling is essential for the development of novel therapeutic approaches. Further investigations are highly recommended to unravel the precise roles of lysosomal ion channels and ion-related signaling in intracellular processes. In conclusion, this review aimed to provide valuable insights into the significance of lysosomal ion channels and ion-related signaling in intracellular signaling and their potential as therapeutic targets. A deeper understanding of the mechanisms involving lysosomal ion channels warrants continued research with the ultimate objective of developing effective therapies for LSDs and other related disorders.
REFERENCES
1. Sterea AM, Almasi S, El Hiani Y. 2018; The hidden potential of lysosomal ion channels: a new era of oncogenes. Cell Calcium. 72:91–103. DOI: 10.1016/j.ceca.2018.02.006. PMID: 29748137.

2. Xiong J, Zhu MX. 2016; Regulation of lysosomal ion homeostasis by channels and transporters. Sci China Life Sci. 59:777–791. DOI: 10.1007/s11427-016-5090-x. PMID: 27430889. PMCID: PMC5147046.

3. Wang H, Zhu Y, Liu H, Liang T, Wei Y. 2023; Advances in drug discovery targeting lysosomal membrane proteins. Pharmaceuticals (Basel). 16:601. DOI: 10.3390/ph16040601. PMID: 37111358. PMCID: PMC10145713. PMID: b51d9efc18bd40f49e2f21ba0254de62.

4. Parenti G, Medina DL, Ballabio A. 2021; The rapidly evolving view of lysosomal storage diseases. EMBO Mol Med. 13:e12836. DOI: 10.15252/emmm.202012836. PMID: 33459519. PMCID: PMC7863408. PMID: af55b965be18440e8fdcf0608db34707.

5. de Duve C, Pressman BC, Gianetto R, Wattiaux R, Appelmans F. 1955; Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J. 60:604–617. DOI: 10.1042/bj0600604. PMID: 13249955. PMCID: PMC1216159.

6. de Duve C. 2005; The lysosome turns fifty. Nat Cell Biol. 7:847–849. DOI: 10.1038/ncb0905-847. PMID: 16136179.

7. Lübke T, Lobel P, Sleat DE. 2009; Proteomics of the lysosome. Biochim Biophys Acta. 1793:625–635. DOI: 10.1016/j.bbamcr.2008.09.018. PMID: 18977398. PMCID: PMC2684028.

8. Yim WW, Mizushima N. 2020; Lysosome biology in autophagy. Cell Discov. 6:6. DOI: 10.1038/s41421-020-0141-7. PMID: 32047650. PMCID: PMC7010707.

9. Tancini B, Buratta S, Delo F, Sagini K, Chiaradia E, Pellegrino RM, Emiliani C, Urbanelli L. 2020; Lysosomal exocytosis: the extracellular role of an intracellular organelle. Membranes (Basel). 10:406. DOI: 10.3390/membranes10120406. PMID: 33316913. PMCID: PMC7764620. PMID: 0936357357dd42f8bbecb9f2943abedb.

10. Bouhamdani N, Comeau D, Turcotte S. 2021; A compendium of information on the lysosome. Front Cell Dev Biol. 9:798262. DOI: 10.3389/fcell.2021.798262. PMID: 34977038. PMCID: PMC8714965. PMID: 988e36504bac4d23b49574ab08014008.

11. Shin HR, Zoncu R. 2020; The lysosome at the intersection of cellular growth and destruction. Dev Cell. 54:226–238. DOI: 10.1016/j.devcel.2020.06.010. PMID: 32610045. PMCID: PMC7959181.

12. Ballabio A, Bonifacino JS. 2020; Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol. 21:101–118. DOI: 10.1038/s41580-019-0185-4. PMID: 31768005.

13. Wie J, Liu Z, Song H, Tropea TF, Yang L, Wang H, Liang Y, Cang C, Aranda K, Lohmann J, Yang J, Lu B, Chen-Plotkin AS, Luk KC, Ren D. 2021; A growth-factor-activated lysosomal K+ channel regulates Parkinson's pathology. Nature. 591:431–437. Erratum in: Nature. 2021;592:E10. DOI: 10.1038/s41586-021-03185-z. PMID: 33505021. PMCID: PMC7979525.

14. Perera RM, Zoncu R. 2016; The lysosome as a regulatory hub. Annu Rev Cell Dev Biol. 32:223–253. DOI: 10.1146/annurev-cellbio-111315-125125. PMID: 27501449. PMCID: PMC9345128.

15. Finkbeiner S. 2020; The autophagy lysosomal pathway and neurodegeneration. Cold Spring Harb Perspect Biol. 12:a033993. DOI: 10.1101/cshperspect.a033993. PMID: 30936119. PMCID: PMC6773515.

16. de Martín Garrido N, Aylett CHS. 2020; Nutrient signaling and lysosome positioning crosstalk through a multifunctional protein, folliculin. Front Cell Dev Biol. 8:108. DOI: 10.3389/fcell.2020.00108. PMID: 32195250. PMCID: PMC7063858. PMID: a207b9a848884d16b361453f2d92c348.

17. Rabanal-Ruiz Y, Korolchuk VI. 2018; mTORC1 and nutrient homeostasis: the central role of the lysosome. Int J Mol Sci. 19:818. DOI: 10.3390/ijms19030818. PMID: 29534520. PMCID: PMC5877679. PMID: 752b601aed9e4a058aacba110a9d833b.

18. Yogalingam G, Luu AR, Prill H, Lo MJ, Yip B, Holtzinger J, Christianson T, Aoyagi-Scharber M, Lawrence R, Crawford BE, LeBowitz JH. 2019; BMN 250, a fusion of lysosomal alpha-N-acetylglucosaminidase with IGF2, exhibits different patterns of cellular uptake into critical cell types of Sanfilippo syndrome B disease pathogenesis. PLoS One. 14:e0207836. DOI: 10.1371/journal.pone.0207836. PMID: 30657762. PMCID: PMC6338363. PMID: 61f67561557e43deb10bf75caf69b046.

19. Jia R, Bonifacino JS. 2019; Lysosome positioning influences mTORC2 and AKT signaling. Mol Cell. 75:26–38.e3. DOI: 10.1016/j.molcel.2019.05.009. PMID: 31130364. PMCID: PMC7446307.

20. Liu B, Palmfeldt J, Lin L, Colaço A, Clemmensen KKB, Huang J, Xu F, Liu X, Maeda K, Luo Y, Jäättelä M. 2018; STAT3 associates with vacuolar H+-ATPase and regulates cytosolic and lysosomal pH. Cell Res. 28:996–1012. DOI: 10.1038/s41422-018-0080-0. PMID: 30127373. PMCID: PMC6170402.

21. Lloyd-Lewis B, Krueger CC, Sargeant TJ, D'Angelo ME, Deery MJ, Feret R, Howard JA, Lilley KS, Watson CJ. 2018; Stat3-mediated alterations in lysosomal membrane protein composition. J Biol Chem. 293:4244–4261. DOI: 10.1074/jbc.RA118.001777. PMID: 29343516. PMCID: PMC5868265.

22. Liu B, Chen R, Zhang Y, Huang J, Luo Y, Rosthøj S, Zhao C, Jäättelä M. 2023; Cationic amphiphilic antihistamines inhibit STAT3 via Ca2+-dependent lysosomal H+ efflux. Cell Rep. 42:112137. DOI: 10.1016/j.celrep.2023.112137. PMID: 36807142. PMCID: PMC9989825.
23. Zhang CS, Jiang B, Li M, Zhu M, Peng Y, Zhang YL, Wu YQ, Li TY, Liang Y, Lu Z, Lian G, Liu Q, Guo H, Yin Z, Ye Z, Han J, Wu JW, Yin H, Lin SY, Lin SC. 2014; The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 20:526–540. DOI: 10.1016/j.cmet.2014.06.014. PMID: 25002183.

24. Wang F, Gómez-Sintes R, Boya P. 2018; Lysosomal membrane permeabilization and cell death. Traffic. 19:918–931. DOI: 10.1111/tra.12613. PMID: 30125440.

25. Er EE, Mendoza MC, Mackey AM, Rameh LE, Blenis J. 2013; AKT facilitates EGFR trafficking and degradation by phosphorylating and activating PIKfyve. Sci Signal. 6:ra45. DOI: 10.1126/scisignal.2004015. PMID: 23757022. PMCID: PMC4041878.

26. Chadha R, Meador-Woodruff JH. 2020; Downregulated AKT-mTOR signaling pathway proteins in dorsolateral prefrontal cortex in Schizophrenia. Neuropsychopharmacology. 45:1059–1067. DOI: 10.1038/s41386-020-0614-2. PMID: 31952070. PMCID: PMC7162985.

27. Hoeffer CA, Klann E. 2010; mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 33:67–75. DOI: 10.1016/j.tins.2009.11.003. PMID: 19963289. PMCID: PMC2821969.

28. Ishizuka Y, Kakiya N, Witters LA, Oshiro N, Shirao T, Nawa H, Takei N. 2013; AMP-activated protein kinase counteracts brain-derived neurotrophic factor-induced mammalian target of rapamycin complex 1 signaling in neurons. J Neurochem. 127:66–77. DOI: 10.1111/jnc.12362. PMID: 23841933.

29. Dalle Pezze P, Ruf S, Sonntag AG, Langelaar-Makkinje M, Hall P, Heberle AM, Razquin Navas P, van Eunen K, Tölle RC, Schwarz JJ, Wiese H, Warscheid B, Deitersen J, Stork B, Fäßler E, Schäuble S, Hahn U, Horvatovich P, Shanley DP, Thedieck K. 2016; A systems study reveals concurrent activation of AMPK and mTOR by amino acids. Nat Commun. 7:13254. DOI: 10.1038/ncomms13254. PMID: 27869123. PMCID: PMC5121333. PMID: 4a285de016fb4d67ada2a43c868dc313.

30. Jeger JL. 2020; Endosomes, lysosomes, and the role of endosomal and lysosomal biogenesis in cancer development. Mol Biol Rep. 47:9801–9810. DOI: 10.1007/s11033-020-05993-4. PMID: 33185829.

31. Reggiori F, Gabius HJ, Aureli M, Römer W, Sonnino S, Eskelinen EL. 2021; Glycans in autophagy, endocytosis and lysosomal functions. Glycoconj J. 38:625–647. DOI: 10.1007/s10719-021-10007-x. PMID: 34390447. PMCID: PMC8497297.

32. Trivedi PC, Bartlett JJ, Pulinilkunnil T. 2020; Lysosomal biology and function: modern view of cellular debris bin. Cells. 9:1131. DOI: 10.3390/cells9051131. PMID: 32375321. PMCID: PMC7290337. PMID: c1e053a9d2a14302ab5abf8ecd22d51c.

33. Bonam SR, Wang F, Muller S. 2019; Lysosomes as a therapeutic target. Nat Rev Drug Discov. 18:923–948. DOI: 10.1038/s41573-019-0036-1. PMID: 31477883. PMCID: PMC7097195.

34. Bajaj L, Lotfi P, Pal R, Ronza AD, Sharma J, Sardiello M. 2019; Lysosome biogenesis in health and disease. J Neurochem. 148:573–589. DOI: 10.1111/jnc.14564. PMID: 30092616. PMCID: PMC6368902.

35. Pfeffer SR. 2019; NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes. J Biol Chem. 294:1706–1709. DOI: 10.1074/jbc.TM118.004165. PMID: 30710017. PMCID: PMC6364775.

36. Altuzar J, Notbohm J, Stein F, Haberkant P, Hempelmann P, Heybrock S, Worsch J, Saftig P, Höglinger D. 2023; Lysosome-targeted multifunctional lipid probes reveal the sterol transporter NPC1 as a sphingosine interactor. Proc Natl Acad Sci U S A. 120:e2213886120. DOI: 10.1073/pnas.2213886120. PMID: 36893262.

37. Henne WM. 2021; How NPC1 loss twists the TORCque of lysosomes. Dev Cell. 56:251–252. DOI: 10.1016/j.devcel.2021.01.007. PMID: 33561418.

38. Patel S, Ramakrishnan L, Rahman T, Hamdoun A, Marchant JS, Taylor CW, Brailoiu E. 2011; The endo-lysosomal system as an NAADP-sensitive acidic Ca(2+) store: role for the two-pore channels. Cell Calcium. 50:157–167. DOI: 10.1016/j.ceca.2011.03.011. PMID: 21529939. PMCID: PMC3160778.

39. Yuan Y, Jaślan D, Rahman T, Bolsover SR, Arige V, Wagner LE 2nd, Abrahamian C, Tang R, Keller M, Hartmann J, Rosato AS, Weiden EM, Bracher F, Yule DI, Grimm C, Patel S. 2022; Segregated cation flux by TPC2 biases Ca2+ signaling through lysosomes. Nat Commun. 13:4481. DOI: 10.1038/s41467-022-31959-0. PMID: 35918320. PMCID: PMC9346130. PMID: db6d7bc0a8014e758a1eca79b88380fb.
40. Lagostena L, Festa M, Pusch M, Carpaneto A. 2017; The human two-pore channel 1 is modulated by cytosolic and luminal calcium. Sci Rep. 7:43900. DOI: 10.1038/srep43900. PMID: 28252105. PMCID: PMC5333365.

41. Calcraft PJ, Ruas M, Pan Z, Cheng X, Arredouani A, Hao X, Tang J, Rietdorf K, Teboul L, Chuang KT, Lin P, Xiao R, Wang C, Zhu Y, Lin Y, Wyatt CN, Parrington J, Ma J, Evans AM, Galione A, et al. 2009; NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature. 459:596–600. DOI: 10.1038/nature08030. PMID: 19387438. PMCID: PMC2761823.

42. Cang C, Zhou Y, Navarro B, Seo YJ, Aranda K, Shi L, Battaglia-Hsu S, Nissim I, Clapham DE, Ren D. 2013; mTOR regulates lysosomal ATP-sensitive two-pore Na(+) channels to adapt to metabolic state. Cell. 152:778–790. DOI: 10.1016/j.cell.2013.01.023. PMID: 23394946. PMCID: PMC3908667.

43. Wang X, Zhang X, Dong XP, Samie M, Li X, Cheng X, Goschka A, Shen D, Zhou Y, Harlow J, Zhu MX, Clapham DE, Ren D, Xu H. 2012; TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell. 151:372–383. DOI: 10.1016/j.cell.2012.08.036. PMID: 23063126. PMCID: PMC3475186.

44. She J, Guo J, Chen Q, Zeng W, Jiang Y, Bai XC. 2018; Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel. Nature. 556:130–134. DOI: 10.1038/nature26139. PMID: 29562233. PMCID: PMC5886804.

45. Patel S, Yuan Y, Gunaratne GS, Rahman T, Marchant JS. 2022; Activation of endo-lysosomal two-pore channels by NAADP and PI(3,5)P2. Five things to know. Cell Calcium. 103:102543. DOI: 10.1016/j.ceca.2022.102543. PMID: 35123238. PMCID: PMC9552313.
46. Russell T, Gangotia D, Barry G. 2022; Assessing the potential of repurposing ion channel inhibitors to treat emerging viral diseases and the role of this host factor in virus replication. Biomed Pharmacother. 156:113850. DOI: 10.1016/j.biopha.2022.113850. PMID: 36411658.

47. Guo J, Zeng W, Jiang Y. 2017; Tuning the ion selectivity of two-pore channels. Proc Natl Acad Sci U S A. 114:1009–1014. DOI: 10.1073/pnas.1616191114. PMID: 28096396. PMCID: PMC5293054.

48. She J, Zeng W, Guo J, Chen Q, Bai XC, Jiang Y. 2019; Structural mechanisms of phospholipid activation of the human TPC2 channel. Elife. 8:e45222. DOI: 10.7554/eLife.45222. PMID: 30860481. PMCID: PMC6424560. PMID: 55bfa42d23844a81bd020fd32b23acbb.

49. Castonguay J, Orth JHC, Müller T, Sleman F, Grimm C, Wahl-Schott C, Biel M, Mallmann RT, Bildl W, Schulte U, Klugbauer N. 2017; The two-pore channel TPC1 is required for efficient protein processing through early and recycling endosomes. Sci Rep. 7:10038. DOI: 10.1038/s41598-017-10607-4. PMID: 28855648. PMCID: PMC5577145.

50. Moccia F, Negri S, Faris P, Perna A, De Luca A, Soda T, Berra-Romani R, Guerra G. 2021; Targeting endolysosomal two-pore channels to treat cardiovascular disorders in the novel COronaVIrus Disease 2019. Front Physiol. 12:629119. Erratum in: Front Physiol. 2021;12:690189. DOI: 10.3389/fphys.2021.690189. PMID: 34122151. PMCID: PMC8195275. PMID: 508aef2cfe2f416387a7d71a4044f88a.

51. Penny CJ, Vassileva K, Jha A, Yuan Y, Chee X, Yates E, Mazzon M, Kilpatrick BS, Muallem S, Marsh M, Rahman T, Patel S. 2019; Mining of Ebola virus entry inhibitors identifies approved drugs as two-pore channel pore blockers. Biochim Biophys Acta Mol Cell Res. 1866:1151–1161. DOI: 10.1016/j.bbamcr.2018.10.022. PMID: 30408544. PMCID: PMC7114365.

52. Heister PM, Poston RN. 2020; Pharmacological hypothesis: TPC2 antagonist tetrandrine as a potential therapeutic agent for COVID-19. Pharmacol Res Perspect. 8:e00653. DOI: 10.1002/prp2.653. PMID: 32930523. PMCID: PMC7503088.

53. Lin PH, Duann P, Komazaki S, Park KH, Li H, Sun M, Sermersheim M, Gumpper K, Parrington J, Galione A, Evans AM, Zhu MX, Ma J. 2015; Lysosomal two-pore channel subtype 2 (TPC2) regulates skeletal muscle autophagic signaling. J Biol Chem. 290:3377–3389. DOI: 10.1074/jbc.M114.608471. PMID: 25480788. PMCID: PMC4319008.

54. Grimm C, Holdt LM, Chen CC, Hassan S, Müller C, Jörs S, Cuny H, Kissing S, Schröder B, Butz E, Northoff B, Castonguay J, Luber CA, Moser M, Spahn S, Lüllmann-Rauch R, Fendel C, Klugbauer N, Griesbeck O, Haas A, et al. 2014; High susceptibility to fatty liver disease in two-pore channel 2-deficient mice. Nat Commun. 5:4699. DOI: 10.1038/ncomms5699. PMID: 25144390.

55. Patel S, Kilpatrick BS. 2018; Two-pore channels and disease. Biochim Biophys Acta Mol Cell Res. 1865(11 Pt B):1678–1686. DOI: 10.1016/j.bbamcr.2018.05.004. PMID: 29746898. PMCID: PMC6162333.

56. Sakurai Y, Kolokoltsov AA, Chen CC, Tidwell MW, Bauta WE, Klugbauer N, Grimm C, Wahl-Schott C, Biel M, Davey RA. 2015; Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science. 347:995–998. DOI: 10.1126/science.1258758. PMID: 25722412. PMCID: PMC4550587.

57. Gerndt S, Krogsaeter E, Patel S, Bracher F, Grimm C. 2020; Discovery of lipophilic two-pore channel agonists. FEBS J. 287:5284–5293. DOI: 10.1111/febs.15432. PMID: 32478984.

58. Chapel A, Kieffer-Jaquinod S, Sagné C, Verdon Q, Ivaldi C, Mellal M, Thirion J, Jadot M, Bruley C, Garin J, Gasnier B, Journet A. 2013; An extended proteome map of the lysosomal membrane reveals novel potential transporters. Mol Cell Proteomics. 12:1572–1588. DOI: 10.1074/mcp.M112.021980. PMID: 23436907. PMCID: PMC3675815.

59. Cang C, Aranda K, Seo YJ, Gasnier B, Ren D. 2015; TMEM175 is an organelle K(+) channel regulating lysosomal function. Cell. 162:1101–1112. DOI: 10.1016/j.cell.2015.08.002. PMID: 26317472.

60. Chia R, Sabir MS, Bandres-Ciga S, Saez-Atienzar S, Reynolds RH, Gustavsson E, Walton RL, Ahmed S, Viollet C, Ding J, Makarious MB, Diez-Fairen M, Portley MK, Shah Z, Abramzon Y, Hernandez DG, Blauwendraat C, Stone DJ, Eicher J, Parkkinen L, et al. 2021; Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat Genet. 53:294–303. DOI: 10.1038/s41588-021-00785-3. PMID: 33589841. PMCID: PMC7946812.
61. Jinn S, Drolet RE, Cramer PE, Wong AH, Toolan DM, Gretzula CA, Voleti B, Vassileva G, Disa J, Tadin-Strapps M, Stone DJ. 2017; TMEM175 deficiency impairs lysosomal and mitochondrial function and increases α-synuclein aggregation. Proc Natl Acad Sci U S A. 114:2389–2394. DOI: 10.1073/pnas.1616332114. PMID: 28193887. PMCID: PMC5338534.

62. Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, Schulte C, Keller MF, Arepalli S, Letson C, Edsall C, Stefansson H, Liu X, Pliner H, Lee JH, Cheng R, et al. International Parkinson's Disease Genomics Consortium (IPDGC). Parkinson's Study Group (PSG) Parkinson's Research: The Organized GENetics Initiative (PROGENI). 23andMe. GenePD. NeuroGenetics Research Consortium (NGRC). Hussman Institute of Human Genomics (HIHG). Ashkenazi Jewish Dataset Investigator. Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE). North American Brain Expression Consortium (NABEC). United Kingdom Brain Expression Consortium (UKBEC). Greek Parkinson's Disease Consortium. Alzheimer Genetic Analysis Group. 2014; Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet. 46:989–993. DOI: 10.1038/ng.3043. PMID: 25064009. PMCID: PMC4146673.

63. Hopfner F, Mueller SH, Szymczak S, Junge O, Tittmann L, May S, Lohmann K, Grallert H, Lieb W, Strauch K, Müller-Nurasyid M, Berger K, Schormair B, Winkelmann J, Mollenhauer B, Trenkwalder C, Maetzler W, Berg D, Kasten M, Klein C, et al. 2020; Rare variants in specific lysosomal genes are associated with Parkinson's disease. Mov Disord. 35:1245–1248. DOI: 10.1002/mds.28037. PMID: 32267580.

64. Brunner JD, Jakob RP, Schulze T, Neldner Y, Moroni A, Thiel G, Maier T, Schenck S. 2020; Structural basis for ion selectivity in TMEM175 K+ channels. Elife. 9:e53683. DOI: 10.7554/eLife.53683. PMID: 32267231. PMCID: PMC7176437. PMID: f8445f40a87949708e12e0e16e894c2a.

65. Iwaki H, Blauwendraat C, Leonard HL, Liu G, Maple-Grødem J, Corvol JC, Pihlstrøm L, van Nimwegen M, Hutten SJ, Nguyen KH, Rick J, Eberly S, Faghri F, Auinger P, Scott KM, Wijeyekoon R, Van Deerlin VM, Hernandez DG, Day-Williams AG, Brice A, et al. 2019; Genetic risk of Parkinson disease and progression: an analysis of 13 longitudinal cohorts. Neurol Genet. 5:e348. Erratum in: Neurol Genet. 2019;5:e354. DOI: 10.1212/NXG.0000000000000348. PMID: 31404238. PMCID: PMC6659137.
66. Ge L, Hoa NT, Wilson Z, Arismendi-Morillo G, Kong XT, Tajhya RB, Beeton C, Jadus MR. 2014; Big Potassium (BK) ion channels in biology, disease and possible targets for cancer immunotherapy. Int Immunopharmacol. 22:427–443. DOI: 10.1016/j.intimp.2014.06.040. PMID: 25027630. PMCID: PMC5472047.

67. Cao Q, Zhong XZ, Zou Y, Zhang Z, Toro L, Dong XP. 2015; BK channels alleviate lysosomal storage diseases by providing positive feedback regulation of lysosomal Ca2+ release. Dev Cell. 33:427–441. DOI: 10.1016/j.devcel.2015.04.010. PMID: 25982675.

68. Clapham DE, Runnels LW, Strübing C. 2001; The TRP ion channel family. Nat Rev Neurosci. 2:387–396. DOI: 10.1038/35077544. PMID: 11389472.

69. Xia Z, Xie L, Li D, Hong X, Qin C. 2023; Gene expression of TRPMLs and its regulation by pathogen stimulation. Gene. 864:147291. DOI: 10.1016/j.gene.2023.147291. PMID: 36813061.

70. Zeevi DA, Frumkin A, Bach G. 2007; TRPML and lysosomal function. Biochim Biophys Acta. 1772:851–858. DOI: 10.1016/j.bbadis.2007.01.004. PMID: 17306511.

71. Falardeau JL, Kennedy JC, Acierno JS Jr, Sun M, Stahl S, Goldin E, Slaugenhaupt SA. 2002; Cloning and characterization of the mouse Mcoln1 gene reveals an alternatively spliced transcript not seen in humans. BMC Genomics. 3:3. DOI: 10.1186/1471-2164-3-3. PMID: 11897010. PMCID: PMC88885. PMID: 11fa92b01b79497a9aa3d689a9531be7.

72. Grimm C, Jörs S, Saldanha SA, Obukhov AG, Pan B, Oshima K, Cuajungco MP, Chase P, Hodder P, Heller S. 2010; Small molecule activators of TRPML3. Chem Biol. 17:135–148. DOI: 10.1016/j.chembiol.2009.12.016. PMID: 20189104. PMCID: PMC2834294.

73. Santoni G, Morelli MB, Amantini C, Nabissi M, Santoni M, Santoni A. 2020; Involvement of the TRPML mucolipin channels in viral infections and anti-viral innate immune responses. Front Immunol. 11:739. DOI: 10.3389/fimmu.2020.00739. PMID: 32425938. PMCID: PMC7212413. PMID: fcea628c9be64115b133a20378e66995.

74. Li M, Zhang WK, Benvin NM, Zhou X, Su D, Li H, Wang S, Michailidis IE, Tong L, Li X, Yang J. 2017; Structural basis of dual Ca2+/pH regulation of the endolysosomal TRPML1 channel. Nat Struct Mol Biol. 24:205–213. DOI: 10.1038/nsmb.3362. PMID: 28112729. PMCID: PMC5336481.

75. Venkatachalam K, Hofmann T, Montell C. 2006; Lysosomal localization of TRPML3 depends on TRPML2 and the mucolipidosis-associated protein TRPML1. J Biol Chem. 281:17517–17527. DOI: 10.1074/jbc.M600807200. PMID: 16606612. PMCID: PMC4196876.

76. Di Paola S, Scotto-Rosato A, Medina DL. 2018; TRPML1: The Ca(2+)retaker of the lysosome. Cell Calcium. 69:112–121. DOI: 10.1016/j.ceca.2017.06.006. PMID: 28689729.
77. Dong XP, Cheng X, Mills E, Delling M, Wang F, Kurz T, Xu H. 2008; The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 455:992–996. DOI: 10.1038/nature07311. PMID: 18794901. PMCID: PMC4301259.

78. Dong XP, Shen D, Wang X, Dawson T, Li X, Zhang Q, Cheng X, Zhang Y, Weisman LS, Delling M, Xu H. 2010; PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat Commun. 1:38. DOI: 10.1038/ncomms1037. PMID: 20802798. PMCID: PMC2928581.

79. Soyombo AA, Tjon-Kon-Sang S, Rbaibi Y, Bashllari E, Bisceglia J, Muallem S, Kiselyov K. 2006; TRP-ML1 regulates lysosomal pH and acidic lysosomal lipid hydrolytic activity. J Biol Chem. 281:7294–7301. DOI: 10.1074/jbc.M508211200. PMID: 16361256.

80. Samie MA, Grimm C, Evans JA, Curcio-Morelli C, Heller S, Slaugenhaupt SA, Cuajungco MP. 2009; The tissue-specific expression of TRPML2 (MCOLN-2) gene is influenced by the presence of TRPML1. Pflugers Arch. 459:79–91. DOI: 10.1007/s00424-009-0716-5. PMID: 19763610. PMCID: PMC2913554.

81. Karacsonyi C, Miguel AS, Puertollano R. 2007; Mucolipin-2 localizes to the Arf6-associated pathway and regulates recycling of GPI-APs. Traffic. 8:1404–1414. DOI: 10.1111/j.1600-0854.2007.00619.x. PMID: 17662026.

82. Sun L, Hua Y, Vergarajauregui S, Diab HI, Puertollano R. 2015; Novel role of TRPML2 in the regulation of the innate immune response. J Immunol. 195:4922–4932. DOI: 10.4049/jimmunol.1500163. PMID: 26432893. PMCID: PMC4637233.

83. Hirschi M, Herzik MA Jr, Wie J, Suo Y, Borschel WF, Ren D, Lander GC, Lee SY. 2017; Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Nature. 550:411–414. DOI: 10.1038/nature24055. PMID: 29019979. PMCID: PMC5762132.

84. Kim HJ, Li Q, Tjon-Kon-Sang S, So I, Kiselyov K, Soyombo AA, Muallem S. 2008; A novel mode of TRPML3 regulation by extracytosolic pH absent in the varitint-waddler phenotype. EMBO J. 27:1197–1205. DOI: 10.1038/emboj.2008.56. PMID: 18369318. PMCID: PMC2367400.

85. Venkatachalam K, Wong CO, Zhu MX. 2015; The role of TRPMLs in endolysosomal trafficking and function. Cell Calcium. 58:48–56. DOI: 10.1016/j.ceca.2014.10.008. PMID: 25465891. PMCID: PMC4412768.

86. Choi S, Kim HJ. 2014; The Ca2+ channel TRPML3 specifically interacts with the mammalian ATG8 homologue GATE16 to regulate autophagy. Biochem Biophys Res Commun. 443:56–61. DOI: 10.1016/j.bbrc.2013.11.044. PMID: 24269818.

87. Kim SW, Kim DH, Park KS, Kim MK, Park YM, Muallem S, So I, Kim HJ. 2019; Palmitoylation controls trafficking of the intracellular Ca2+ channel MCOLN3/TRPML3 to regulate autophagy. Autophagy. 15:327–340. DOI: 10.1080/15548627.2018.1518671. PMID: 30215288. PMCID: PMC6333453.

88. Kim HJ, Soyombo AA, Tjon-Kon-Sang S, So I, Muallem S. 2009; The Ca(2+) channel TRPML3 regulates membrane trafficking and autophagy. Traffic. 10:1157–1167. DOI: 10.1111/j.1600-0854.2009.00924.x. PMID: 19522758. PMCID: PMC2993507.
89. Grimm C, Cuajungco MP, van Aken AF, Schnee M, Jörs S, Kros CJ, Ricci AJ, Heller S. 2007; A helix-breaking mutation in TRPML3 leads to constitutive activity underlying deafness in the varitint-waddler mouse. Proc Natl Acad Sci U S A. 104:19583–19588. DOI: 10.1073/pnas.0709846104. PMID: 18048323. PMCID: PMC2148332.

90. Jentsch TJ, Pusch M. 2018; CLC chloride channels and transporters: structure, function, physiology, and disease. Physiol Rev. 98:1493–1590. DOI: 10.1152/physrev.00047.2017. PMID: 29845874.

91. Zifarelli G, Pusch M. 2010; CLC transport proteins in plants. FEBS Lett. 584:2122–2127. DOI: 10.1016/j.febslet.2009.12.042. PMID: 20036660.

92. Kornak U, Kasper D, Bösl MR, Kaiser E, Schweizer M, Schulz A, Friedrich W, Delling G, Jentsch TJ. 2001; Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell. 104:205–215. DOI: 10.1016/S0092-8674(01)00206-9. PMID: 11207362.

93. Kasper D, Planells-Cases R, Fuhrmann JC, Scheel O, Zeitz O, Ruether K, Schmitt A, Poët M, Steinfeld R, Schweizer M, Kornak U, Jentsch TJ. 2005; Loss of the chloride channel ClC-7 leads to lysosomal storage disease and neurodegeneration. EMBO J. 24:1079–1091. DOI: 10.1038/sj.emboj.7600576. PMID: 15706348. PMCID: PMC554126.

94. Brandt S, Jentsch TJ. 1995; ClC-6 and ClC-7 are two novel broadly expressed members of the CLC chloride channel family. FEBS Lett. 377:15–20. DOI: 10.1016/0014-5793(95)01298-2. PMID: 8543009.

95. Stauber T, Weinert S, Jentsch TJ. 2012; Cell biology and physiology of CLC chloride channels and transporters. Compr Physiol. 2:1701–1744. DOI: 10.1002/cphy.c110038. PMID: 23723021.

96. Bergsdorf EY, Zdebik AA, Jentsch TJ. 2009; Residues important for nitrate/proton coupling in plant and mammalian CLC transporters. J Biol Chem. 284:11184–11193. DOI: 10.1074/jbc.M901170200. PMID: 19261613. PMCID: PMC2670123.

97. Schrecker M, Korobenko J, Hite RK. 2020; Cryo-EM structure of the lysosomal chloride-proton exchanger CLC-7 in complex with OSTM1. Elife. 9:e59555. DOI: 10.7554/eLife.59555. PMID: 32749217. PMCID: PMC7440919. PMID: 0d3b68ed2f11477cbed44d09d2053b47.

98. Lange PF, Wartosch L, Jentsch TJ, Fuhrmann JC. 2006; ClC-7 requires Ostm1 as a beta-subunit to support bone resorption and lysosomal function. Nature. 440:220–223. DOI: 10.1038/nature04535. PMID: 16525474.

99. Jentsch TJ. 2007; Chloride and the endosomal-lysosomal pathway: emerging roles of CLC chloride transporters. J Physiol. 578:633–640. DOI: 10.1113/jphysiol.2006.124719. PMID: 17110406. PMCID: PMC2151350.

100. Chalhoub N, Benachenhou N, Rajapurohitam V, Pata M, Ferron M, Frattini A, Villa A, Vacher J. 2003; Grey-lethal mutation induces severe malignant autosomal recessive osteopetrosis in mouse and human. Nat Med. 9:399–406. DOI: 10.1038/nm842. PMID: 12627228.

101. Leisle L, Ludwig CF, Wagner FA, Jentsch TJ, Stauber T. 2011; ClC-7 is a slowly voltage-gated 2Cl(-)/1H(+)-exchanger and requires Ostm1 for transport activity. EMBO J. 30:2140–2152. DOI: 10.1038/emboj.2011.137. PMID: 21527911. PMCID: PMC3117652.
102. Wu JZ, Zeziulia M, Kwon W, Jentsch TJ, Grinstein S, Freeman SA. 2023; ClC-7 drives intraphagosomal chloride accumulation to support hydrolase activity and phagosome resolution. J Cell Biol. 222:e202208155. DOI: 10.1083/jcb.202208155. PMID: 37010469.

103. Leray X, Hilton JK, Nwangwu K, Becerril A, Mikusevic V, Fitzgerald G, Amin A, Weston MR, Mindell JA. 2022; Tonic inhibition of the chloride/proton antiporter ClC-7 by PI(3,5)P2 is crucial for lysosomal pH maintenance. Elife. 11:e74136. DOI: 10.7554/eLife.74136. PMID: 35670560. PMCID: PMC9242644. PMID: ba99f83a0c944e9299218d4891a98871.

104. Klaus F, Laufer J, Czarkowski K, Strutz-Seebohm N, Seebohm G, Lang F. 2009; PIKfyve-dependent regulation of the Cl- channel ClC-2. Biochem Biophys Res Commun. 381:407–411. DOI: 10.1016/j.bbrc.2009.02.053. PMID: 19232516.

105. Zifarelli G. 2022; The role of the lysosomal Cl-/H+ antiporter ClC-7 in osteopetrosis and neurodegeneration. Cells. 11:366. DOI: 10.3390/cells11030366. PMID: 35159175. PMCID: PMC8833911. PMID: b2d3c6eaa5074516a368f8703a06517c.

106. Kroemer G, Jäättelä M. 2005; Lysosomes and autophagy in cell death control. Nat Rev Cancer. 5:886–897. DOI: 10.1038/nrc1738. PMID: 16239905.

107. Hwang J, Estick CM, Ikonne US, Butler D, Pait MC, Elliott LH, Ruiz S, Smith K, Rentschler KM, Mundell C, Almeida MF, Stumbling Bear N, Locklear JP, Abumohsen Y, Ivey CM, Farizatto KLG, Bahr BA. 2019; The role of lysosomes in a broad disease-modifying approach evaluated across transgenic mouse models of Alzheimer's disease and Parkinson's disease and models of mild cognitive impairment. Int J Mol Sci. 20:4432. DOI: 10.3390/ijms20184432. PMID: 31505809. PMCID: PMC6770842. PMID: 55c4732172c64b61960ce84245284300.

108. Navarro-Romero A, Montpeyó M, Martinez-Vicente M. 2020; The emerging role of the lysosome in Parkinson's disease. Cells. 9:2399. DOI: 10.3390/cells9112399. PMID: 33147750. PMCID: PMC7692401. PMID: f00f10a0147e479b9582e2cca8ef53bc.

109. Wiwatpanit T, Remis NN, Ahmad A, Zhou Y, Clancy JC, Cheatham MA, García-Añoveros J. 2018; Codeficiency of lysosomal mucolipins 3 and 1 in cochlear hair cells diminishes outer hair cell longevity and accelerates age-related hearing loss. J Neurosci. 38:3177–3189. DOI: 10.1523/JNEUROSCI.3368-17.2018. PMID: 29453205. PMCID: PMC5884457.

110. Hayashi K, Suzuki Y, Fujimoto C, Kanzaki S. 2020; Molecular mechanisms and biological functions of autophagy for genetics of hearing impairment. Genes (Basel). 11:1331. DOI: 10.3390/genes11111331. PMID: 33187328. PMCID: PMC7697636. PMID: 52524255ba0f479bb50e7f3c7d4a5c63.

111. Mizunoe Y, Kobayashi M, Tagawa R, Nakagawa Y, Shimano H, Higami Y. 2019; Association between lysosomal dysfunction and obesity-related pathology: a key knowledge to prevent metabolic syndrome. Int J Mol Sci. 20:3688. DOI: 10.3390/ijms20153688. PMID: 31357643. PMCID: PMC6696452. PMID: 4581e64a690e4be8b3449a6658deb4af.

112. Rawnsley DR, Diwan A. 2020; Lysosome impairment as a trigger for inflammation in obesity: the proof is in the fat. EBioMedicine. 56:102824. DOI: 10.1016/j.ebiom.2020.102824. PMID: 32540774. PMCID: PMC7300142.

113. Barnett BS, Ziegler K, Doblin R, Carlo AD. 2022; Is psychedelic use associated with cancer?: interrogating a half-century-old claim using contemporary population-level data. J Psychopharmacol. 36:1118–1128. DOI: 10.1177/02698811221117536. PMID: 35971893.

114. Ng PY, Ribet ABP, Guo Q, Mullin BH, Tan JWY, Landao-Bassonga E, Stephens S, Chen K, Yuan J, Abudulai L, Bollen M, Nguyen ETTT, Kular J, Papadimitriou JM, Søe K, Teasdale RD, Xu J, Parton RG, Takayanagi H, Pavlos NJ. 2023; Sugar transporter Slc37a2 regulates bone metabolism in mice via a tubular lysosomal network in osteoclasts. Nat Commun. 14:906. DOI: 10.1038/s41467-023-36484-2. PMID: 36810735. PMCID: PMC9945426. PMID: cfbfda3751ad47868d62c17e221301bd.

115. Larsen LE, van den Boogert MAW, Rios-Ocampo WA, Jansen JC, Conlon D, Chong PLE, Levels JHM, Eilers RE, Sachdev VV, Zelcer N, Raabe T, He M, Hand NJ, Drenth JPH, Rader DJ, Stroes ESG, Lefeber DJ, Jonker JW, Holleboom AG. 2022; Defective lipid droplet-lysosome interaction causes fatty liver disease as evidenced by human mutations in TMEM199 and CCDC115. Cell Mol Gastroenterol Hepatol. 13:583–597. DOI: 10.1016/j.jcmgh.2021.09.013. PMID: 34626841. PMCID: PMC8688563.

116. Machado ER, Annunziata I, van de Vlekkert D, Grosveld GC. d'Azzo A. 2021; Lysosomes and cancer progression: a malignant liaison. Front Cell Dev Biol. 9:642494. DOI: 10.3389/fcell.2021.642494. PMID: 33718382. PMCID: PMC7952443. PMID: 1989c67ff27d4918bc2104fec8e3962f.

117. Zhao Z, Qin P, Huang YW. 2021; Lysosomal ion channels involved in cellular entry and uncoating of enveloped viruses: implications for therapeutic strategies against SARS-CoV-2. Cell Calcium. 94:102360. DOI: 10.1016/j.ceca.2021.102360. PMID: 33516131. PMCID: PMC7825922.

118. Wang X, Melino G, Shi Y. 2021; Actively or passively deacidified lysosomes push β-coronavirus egress. Cell Death Dis. 12:235. DOI: 10.1038/s41419-021-03501-5. PMID: 33664221. PMCID: PMC7930523. PMID: cf10ada99fb64de9b7aba8cfa3f4fc60.

119. Vardi A, Pri-Or A, Wigoda N, Grishchuk Y, Futerman AH. 2021; Proteomics analysis of a human brain sample from a mucolipidosis type IV patient reveals pathophysiological pathways. Orphanet J Rare Dis. 16:39. DOI: 10.1186/s13023-021-01679-7. PMID: 33478506. PMCID: PMC7818904. PMID: 67440a526e7946dd9a8095ed8498df40.

120. Vacca F, Vossio S, Mercier V, Moreau D, Johnson S, Scott CC, Montoya JP, Moniatte M, Gruenberg J. 2019; Cyclodextrin triggers MCOLN1-dependent endo-lysosome secretion in Niemann-Pick type C cells. J Lipid Res. 60:832–843. DOI: 10.1194/jlr.M089979. PMID: 30709900. PMCID: PMC6446697.

121. Gibson PG, Qin L, Puah SH. 2020; COVID-19 acute respiratory distress syndrome (ARDS): clinical features and differences from typical pre-COVID-19 ARDS. Med J Aust. 213:54–56.e1. DOI: 10.5694/mja2.50674. PMID: 32572965. PMCID: PMC7361309.

122. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, et al. 2020; Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 395:497–506. Erratum in: Lancet. 2020 Jan 30. DOI: 10.1016/S0140-6736(20)30183-5. PMID: 31986264.

123. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, et al. 2020; A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579:270–273. Erratum in: Nature. 2020;588:E6. DOI: 10.1038/s41586-020-2951-z. PMID: 33199918. PMCID: PMC9744119.

124. Nishiga M, Wang DW, Han Y, Lewis DB, Wu JC. 2020; COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives. Nat Rev Cardiol. 17:543–558. DOI: 10.1038/s41569-020-0413-9. PMID: 32690910. PMCID: PMC7370876.

125. Zheng YY, Ma YT, Zhang JY, Xie X. 2020; COVID-19 and the cardiovascular system. Nat Rev Cardiol. 17:259–260. DOI: 10.1038/s41569-020-0360-5. PMID: 32139904. PMCID: PMC7095524.

126. Grimm C, Tang R. 2020; Could an endo-lysosomal ion channel be the Achilles heel of SARS-CoV2? Cell Calcium. 88:102212. DOI: 10.1016/j.ceca.2020.102212. PMID: 32402856. PMCID: PMC7201244.

127. Arlt E, Fraticelli M, Tsvilovskyy V, Nadolni W, Breit A, O'Neill TJ, Resenberger S, Wennemuth G, Wahl-Schott C, Biel M, Grimm C, Freichel M, Gudermann T, Klugbauer N, Boekhoff I, Zierler S. 2020; TPC1 deficiency or blockade augments systemic anaphylaxis and mast cell activity. Proc Natl Acad Sci U S A. 117:18068–18078. DOI: 10.1073/pnas.1920122117. PMID: 32661165. PMCID: PMC7395440.

128. Scotto Rosato A, Krogsaeter EK, Jaślan D, Abrahamian C, Montefusco S, Soldati C, Spix B, Pizzo MT, Grieco G, Böck J, Wyatt A, Wünkhaus D, Passon M, Stieglitz M, Keller M, Hermey G, Markmann S, Gruber-Schoffnegger D, Cotman S, Johannes L, et al. 2022; TPC2 rescues lysosomal storage in mucolipidosis type IV, Niemann-Pick type C1, and Batten disease. EMBO Mol Med. 14:e15377. DOI: 10.15252/emmm.202115377. PMID: 35929194. PMCID: PMC9449600. PMID: 81fb1739060044f693962704d333fab5.

129. Du X, Carvalho-de-Souza JL, Wei C, Carrasquel-Ursulaez W, Lorenzo Y, Gonzalez N, Kubota T, Staisch J, Hain T, Petrossian N, Xu M, Latorre R, Bezanilla F, Gomez CM. 2020; Loss-of-function BK channel mutation causes impaired mitochondria and progressive cerebellar ataxia. Proc Natl Acad Sci U S A. 117:6023–6034. DOI: 10.1073/pnas.1920008117. PMID: 32132200. PMCID: PMC7084159.

130. González-Sanabria N, Echeverría F, Segura I, Alvarado-Sánchez R, Latorre R. 2021; BK in double-membrane organelles: a biophysical, pharmacological, and functional survey. Front Physiol. 12:761474. DOI: 10.3389/fphys.2021.761474. PMID: 34764886. PMCID: PMC8577798. PMID: 916f72ac382d4bec89c04f172357bb28.

131. Park SM, Roache CE, Iffland PH 2nd, Moldenhauer HJ, Matychak KK, Plante AE, Lieberman AG, Crino PB, Meredith A. 2022; BK channel properties correlate with neurobehavioral severity in three KCNMA1-linked channelopathy mouse models. Elife. 11:e77953. DOI: 10.7554/eLife.77953. PMID: 35819138. PMCID: PMC9275823. PMID: 987d4544913248bc9d9308fe770b414a.

132. Caldeira da Silva CC, Cerqueira FM, Barbosa LF, Medeiros MH, Kowaltowski AJ. 2008; Mild mitochondrial uncoupling in mice affects energy metabolism, redox balance and longevity. Aging Cell. 7:552–560. DOI: 10.1111/j.1474-9726.2008.00407.x. PMID: 18505478.

133. Donida B, Marchetti DP, Biancini GB, Deon M, Manini PR, da Rosa HT, Moura DJ, Saffi J, Bender F, Burin MG, Coitinho AS, Giugliani R, Vargas CR. 2015; Oxidative stress and inflammation in mucopolysaccharidosis type IVA patients treated with enzyme replacement therapy. Biochim Biophys Acta. 1852:1012–1019. DOI: 10.1016/j.bbadis.2015.02.004. PMID: 25701642.

134. Teixeira CA, Miranda CO, Sousa VF, Santos TE, Malheiro AR, Solomon M, Maegawa GH, Brites P, Sousa MM. 2014; Early axonal loss accompanied by impaired endocytosis, abnormal axonal transport, and decreased microtubule stability occur in the model of Krabbe's disease. Neurobiol Dis. 66:92–103. DOI: 10.1016/j.nbd.2014.02.012. PMID: 24607884. PMCID: PMC4307018.

135. Eldar-Finkelman H, Martinez A. 2011; GSK-3 inhibitors: preclinical and clinical focus on CNS. Front Mol Neurosci. 4:32. DOI: 10.3389/fnmol.2011.00032. PMID: 22065134. PMCID: PMC3204427. PMID: 2d8a6b266637443d844fbe88e8f844e6.

136. Stepien KM, Cufflin N, Donald A, Jones S, Church H, Hargreaves IP. 2022; Secondary mitochondrial dysfunction as a cause of neurodegenerative dysfunction in lysosomal storage diseases and an overview of potential therapies. Int J Mol Sci. 23:10573. DOI: 10.3390/ijms231810573. PMID: 36142486. PMCID: PMC9503973. PMID: 67a11dedb1d94181b25d0f078f18df87.

137. Zhang X, Chen W, Gao Q, Yang J, Yan X, Zhao H, Su L, Yang M, Gao C, Yao Y, Inoki K, Li D, Shao R, Wang S, Sahoo N, Kudo F, Eguchi T, Ruan B, Xu H. 2019; Rapamycin directly activates lysosomal mucolipin TRP channels independent of mTOR. PLoS Biol. 17:e3000252. DOI: 10.1371/journal.pbio.3000252. PMID: 31112550. PMCID: PMC6528971. PMID: 7572bdcdbb8b4629856919a744d68590.

138. Hooper C, Killick R, Lovestone S. 2008; The GSK3 hypothesis of Alzheimer's disease. J Neurochem. 104:1433–1439. DOI: 10.1111/j.1471-4159.2007.05194.x. PMID: 18088381. PMCID: PMC3073119.

Fig. 1
Schematic lysosomal ion channels and transporters.
The ion movements belong to ion channel directions. Lysosomal ion channels characterized permeability; TRPMLs (few divalent cations, and nonselective cations: yellow), TMEM175, BK (potassium ion: blue), ClC-7 (chloride and hydrogen: green), V-ATPase (hydrogen: black), and TPCs (sodium, and calcium: purple). Compared to the cytosol, lysosomal lumen has high concentrations of Na+, Ca2+, and H+ but not K+. BK, calcium-activated large-conductance K+ channel; TPC, two-pore channel.
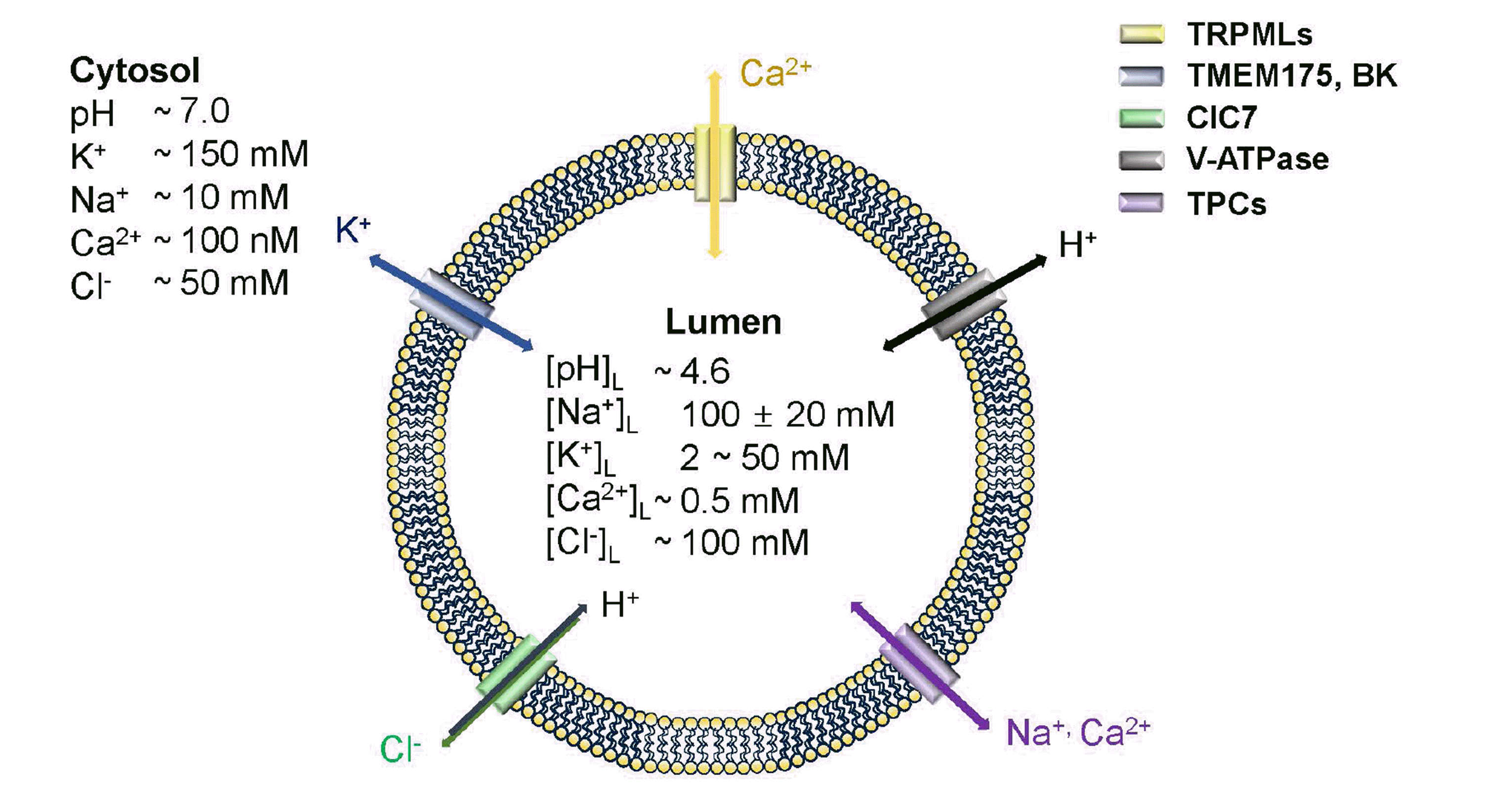
Fig. 2
Interaction between ion channels and regulators.
Amino acids and growth factors bind to receptors on the plasma membrane. PI3kinases increase the concentration of PIP3 from PtdIns(4,5)P2 (PIP2), which binds to the PH domain of AKT. This binding induces a con-formational change that affects downstream signaling pathways such as PIKfyve or mTORC1. AKT phosphorylation occurs at an important residue in the kinase domain (S308) by PDK1 and another residue (S473) in the C-terminal regulatory domain. Phosphorylation of AKT induces changes in PIKfyve, which converts PI(3)P2 to PI(3,5)P2 [25]. BK and TRPML1 channels are promoted by PI(3,5)P2 and reactive oxidative stress (ROS). AKT phosphorylation also inhibits TSC1/2, abolishing Rheb-induced phosphorylation of mTORC1 at residues S2481 and S2448 [26,27]. AMPK signaling is affected by glucose (activation) and setrin (inhibition), AMPK inhibits the mTORC1 [28,29]. TPCs are inhibited by mTORC1 when its domain that interacts with TPC is closed. V-ATPase regulated by mTOR1 through a regulator complex when binding with Rheb. TMEM175 activation requires aa conformational change in PH domain of AKT but does not associate AKT downstream pathways. mTORC1, mechanistic targets of rapamycin complex 1; AMPK, AMP-activated kinase; TPC, two-pore channel; BK, calcium-activated large-conductance K+ channel; TRPML, transient receptor potential mucolipin.
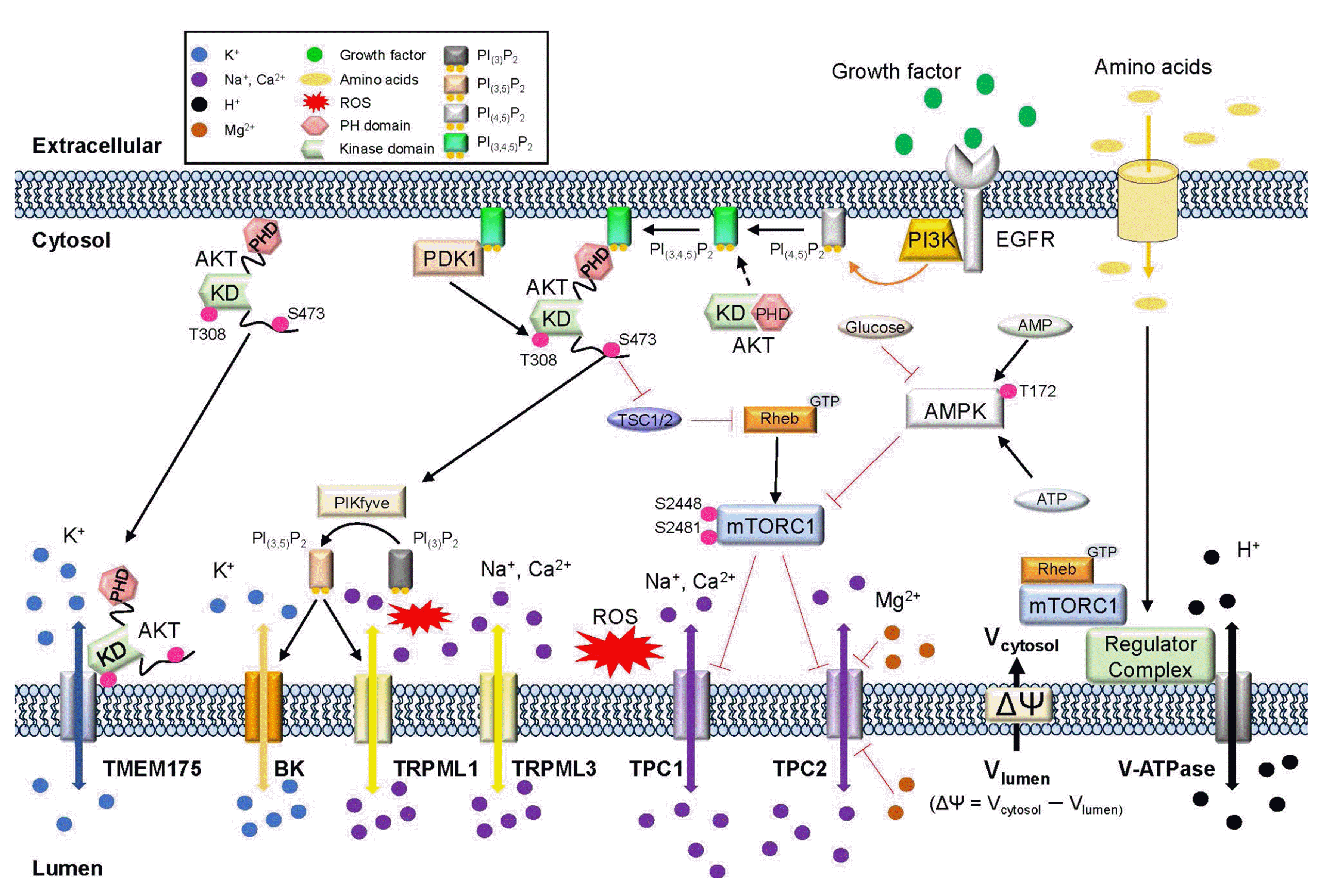
Table 1
Lysosomal ion channels and diseases
| Ion channel name | Transported ion(s) | Disease type |
|---|---|---|
| TPC1 | Na+, Ca2+ | Associated with the fusion of virus [50,120], acute respiratory distress syndrome [115-117] |
| TPC2 | Na+, Ca2+ | Associated with the fusion of virus [50,120], Mucolipidosis type IV (MLIV) [113] |
| TMEM175 | K+ | Parkinson’s disease [13,64] |
| BK | K+ | Cancer [123], seizure [125] |
| TRPML1 |
Na+, Ca2+, Zn+, Fe2+ Cations (nonselective) |
MLIV [113], Niemann–Pick disease [114], Hearing loss [103,104] |
| TRPML2 |
Na+, Ca2+, Zn+, Fe2+ Cations (nonselective) |
|
| TRPML3 |
Na+, Ca2+, Zn+, Fe2+ Cations (nonselective) |
Hearing loss [85,103,104] |
| ClC7 | Cl- |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download