INTRODUCTION
Intraductal papillary mucinous neoplasms (IPMN) is a known precursor to pancreatic ductal adenocarcinoma, the third leading cause of cancer death with an overall five-year survival of 10% [
1]. It has been estimated that up to 49.1% of individuals who have undergone cross-sectional imaging have an incidentally detected pancreatic cystic lesion [
2]. The prevalence of IPMNs is increasing. This increase is likely due to increased incidental detection with expanded use of diagnostic cross-sectional imaging [
2,
3].
The only treatment for IPMN lesions is surgical resection, which carries significant risks of morbidity and mortality [
4]. Ideally, perfect management of IPMNs primarily serves to prevent malignant progression while avoiding unnecessary surgery where possible. Unfortunately, current standard clinical guidelines remain imperfect for distinguishing low-risk IPMNs from high-risk lesions that warrant surgical resection, where high-risk is defined by the presence of high-grade dysplasia or early invasive adenocarcinoma on pathology.
Currently, there are four leading sets of clinical guidelines that dictate management of IPMNs: the 2017 Revised International Consensus Guidelines (widely known as the Fukuoka guidelines) [
5] the 2015 American Gastroenterological Association (AGA) guidelines [
6], the 2018 American College of Gastroenterology (ACG) guidelines [
7], and the 2013 European guidelines published by the European Study Group on Cystic Tumors of the Pancreas [
8]. These guidelines incorporate radiographic criteria, patient signs and symptoms, and serological markers. Studies applying these guidelines retrospectively have found that they are imperfect in distinguishing low-risk from high-risk disease. All four guidelines appear to lead to surgical overtreatment of IPMNs based on histopathologic outcomes [
9]. A recent study analyzed a group of patients who had undergone resection of IPMN and compared their initial indications for surgery with the final histopathologic outcome. It estimated that surgery was justified in only 54%, 59%, and 53% based on Fukuoka, AGA, and European guidelines, respectively [
3,
9].
Studies have also assessed performances of these guidelines in predicting high-risk lesions. Fukuoka guidelines show a sensitivity of 55.6% and a specificity of 73%. AGA guidelines have a sensitivity of 62% and a specificity of 79% [
2,
9-
11] Fukuoka guidelines and European guidelines rarely miss high-risk lesions when they are strictly applied [
9,
10]. However, as previously stated, these guidelines can lead to unnecessary operations and potential overtreatment. Fukuoka guidelines result in unnecessary surgery in up to 30% of patients who ultimately have low-grade lesions. AGA guidelines are more conservative than Fukuoka and European guidelines. However, theAGA guidelines still lead to unnecessary operations in 41.5% of cases [
9]. Moreover, it has been estimated that the AGA criteria can miss up to 45% of high-risk lesions [
9-
11].
Artificial intelligence-based algorithms have been utilized for early detection and prognostication of other cancers including breast, lung, and prostate cancers [
12-
15]. Given the increasing prevalence of IPMNs and morbidity of surgical resection, we aimed to build a machine-learning tool to help identify patients with low-risk IPMNs who might be able to reliably avoid unnecessary surgical resection.
Go to :

MATERIALS AND METHODS
Patient selection
This study utilized a prospectively maintained surgical registry of patients who had undergone resection of an IPMN. A total of 575 patients were identified from January 1, 2000 to January 1, 2018 in a prospectively maintained database of a single institution. This time period was chosen to allow for at least a 3-year follow-up period. All patients were included in this study. Our study was approved by the Partners Institutional Review Board (IRB#: 2002P000153).
Patient characteristics
We collected the following data for each patient: age, gender, ethnicity, family history of cancer, personal history of cancer, abdominal pain, diabetes, steatorrhea, thrombophlebitis, jaundice, history of pancreatitis, and serum carbohydrate antigen (CA) 19-9. We also collected the following data on the IPMNs: multicystic lesions, septations, nodules, cyst location, cyst size, associated pancreatic duct dilation, and final histopathologic outcome.
Machine learning algorithm
A linear support vector machine (SVM) machine learning technique was applied to the extracted patient and IPMN characteristics summarized above. Patient characteristics such as gender, age, ethnicity, family history of any type of cancer, personal history of cancer, jaundice, history of pancreatitis, abdominal pain, history of diabetes, steatorrhea, and thrombophlebitis were included in the model. IPMN characteristics such as multicystic lesions, septations, presence of nodules, tumor location, pancreatic duct dilation, and cyst size were included in the model. Serum CA19-9 was also included in the model. Data were divided into a training/validation set and an independent testing set at a ratio of 4 : 1 (
Fig. 1). Extracted patient and IPMN characteristics were used to train and test a linear SVM-based machine learning model named IPMN-LEARN to predict final pathologic low-grade following IPMN resection. The classifier was optimized using Bayesian optimization. The model was tested using a patient group that was excluded from the training step.
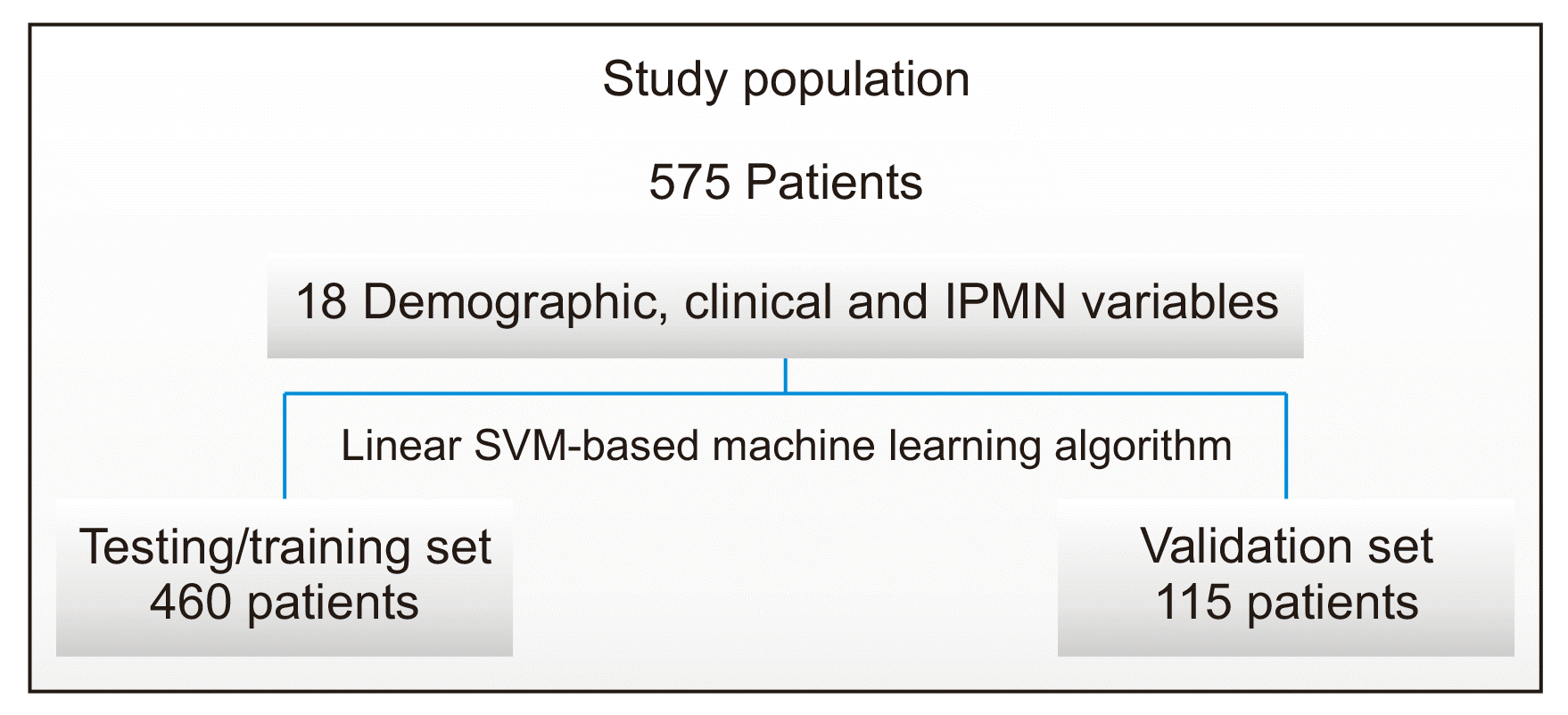 | Fig. 1Data analysis scheme used to predict IPMN grade. A total of 575 patients were enrolled in the study. Data were divided into a training/testing set that included 460 patients and an independent validation set that included 115 patients. Eighteen variables including demographic patient characteristics, clinical information, and IPMN imaging descriptors were used to build a linear SVM-based machine learning model to predict low-grade IPMN status post-surgical resection in this patient population. IPMN, intraductal papillary mucinous neoplasm; SVM, support vector machine. 
|
Model performance assessment
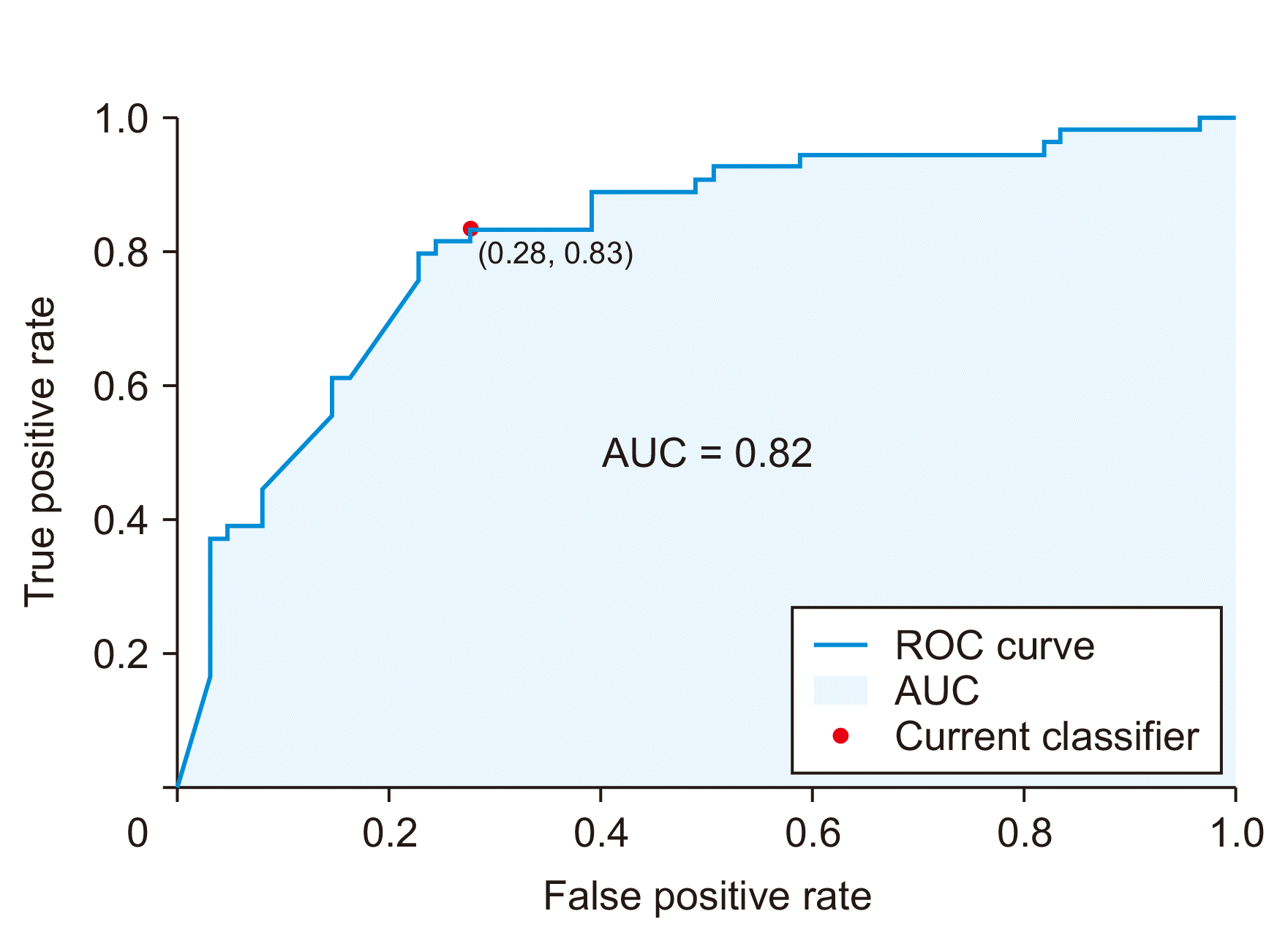
Model performance for IPMN grade prediction following resection was assessed using ROC curve and area under the curve (AUC) analysis. Confusion matrix analysis was performed to determine model accuracy, sensitivity, specificity, positive predictive value, and negative predictive value.
Go to :

DISCUSSION
Our results indicated that IPMN-LEARN, a machine learning algorithm incorporating demographic information, clinical characteristics, and imaging descriptors in patients with IPMN, could effectively predict low-grade disease as determined on pathology following surgical resection. The model that was trained and tested resulted in high sensitivity, high specificity, and high positive predictive value using an independent set of patients who were intentionally excluded from the original model design.
Existing guidelines (Fukuoka, AGA, and European) have been assessed according to their ability to predict a high-risk disease (high-grade dysplasia or adenocarcinoma). Our model, on the other hand, was assessed according to its ability to predict a low-risk disease as defined by the Baltimore consensus meeting for neoplastic precursor lesions in the pancreas [
17]. Although a parallel comparison of performance metrics could be made, our machine learning algorithm provided a higher sensitivity than existing guidelines with similar specificity. Fukuoka guidelines have an estimated sensitivity of 55% and a specificity of 73% and AGA guidelines have a sensitivity of 62% and a specificity of 79% for identifying a high-risk disease [
2,
9-
11]. In contrast, our model had a sensitivity of 83% and a specificity of 72% for predicting a low-risk disease.
Artificial intelligence has a wide range of current and potential applications in the medical sector. For instance, artificial intelligence-based algorithms have been utilized for early detection and prognostication of cancers including breast, lung, and prostate cancers [
12-
15]. Artificial intelligence has been utilized for improved diagnostics, management of disease, and prediction of survival in various cancers. For example, a 2019 study by Rodriguez-Ruiz et al. [
15] has analyzed the performance of a commercially available artificial intelligence system designed to analyze mammograms. The performance of the artificial intelligence system was found to be equivalent to that of radiologists, reinforcing the utility of the platform in a clinical setting [
15]. In another study by Yu et al. [
14], the authors developed a machine learning algorithm that was effective in differentiating between adenocarcinoma and squamous cell carcinoma of the lung as well as predicting survival in patients with non-small cell lung cancer. Moreover, their study found that computers were able to provide insight into specificity of the disease based on histopathology images [
14]. Moving forward, computers might be able to define subtypes of squamous cell carcinoma and adenocarcinoma, paving the way for clinical trials that focus on treatments for subgroups of patients identified by automated analysis of histopathology images. Lastly, in a recent study by Jović et al. [
13], the authors found that several machine learning techniques could reliably predict survival in prostate cancer.
Several recent studies have explored applications of artificial intelligence in pancreatic cancer and its precursors. A review by Dalal et al. [
18] in 2020 discussed the potential benefit and current limitations of artificial intelligence in the detection and management of pancreatic cystic lesions. Their review recognized the significant potential value of radiomics, including high-throughput data extracted from standard images using data-characterization algorithms, allowing for identification of features that might not otherwise be identified by naked human eyes. The authors also highlighted many current limitations, ranging from the challenge of overfitting to the lack of standardization in imaging acquisition and feature analysis across medical centers, which could produce artifacts not attributable to the underlying pathology. In a separate report, Barua et al. [
19] have developed a computational prediction model encapsulating spatial cellular interactions in surgically resected IPMNs that play a role in the transformation of low-grade IPMN cysts to high-grade cysts, which could be used in the future as a risk assessment tool for patients diagnosed with IPMNs. In their study, the authors modeled spatial distance between epithelial cells and immune cells (CD3+CD4+ T cells, CD3+CD8+ T cells, CD68+ macrophages, and PDL1/1+ cells) to determine cyst grade. Their findings highlighted that spatial proximity of cytotoxic T cells to epithelial cells and PD-L1+ macrophages could predict dysplastic grade of the cyst. Additionally, spatial interactions identified in such a study might guide future research concerning the tumor microenvironment [
19]. Finally, Springer et al. [
20] have developed a machine learning algorithm called CompCyst to classify patients with IPMNs as those who require surgery, who should be routinely monitored, and who do not require further surveillance. In their model, they incorporated patient characteristics, imaging findings, and molecular features and generated a prediction model that was more accurate than management dictated by existing clinical and imaging criteria alone [
20].
Our study provides a novel element to prior work utilizing machine learning technology described above by specifically attempting to identify cysts that are less likely to require resection. Given that surgical resection of IPMNs is associated with 40% morbidity and an approximate 2% mortality and that a significant number of patients are found to have an IPMN on imaging [
2], better stratification of patients who are likely to benefit from surgical resection is paramount. The tool developed in this study, IPMN-LEARN, a linear SVM-based machine learning model, might be used as a complement to existing clinical guidelines to better determine which patients diagnosed with an IPMN may benefit from observation vs. surgical resection, thereby contributing to an overall reduction in morbidity and mortality. We chose to develop a model to help predict low-grade dysplasia versus high-grade dysplasia in this study because international pancreatology guidelines used in our center had a high specificity with a quite low risk of missing a cancer. However, we observed approximately 50% of patients who underwent surgery for low-grade dysplasia as their highest grade of dysplasia on histopathological review. Although patients did well long-term following surgery in general, they were exposed to significant potential morbidity and mortality. The goal of this model is to help provide reassurance to patients who are at higher risk of being surgical candidates that they could be managed with imaging surveillance instead of immediate surgery. Using this model, it is not possible to ascertain which specific variables carry the most “weight” in determining low-risk potential. Future models can be developed to better identify which clinical and imaging characteristics are associated with low-risk for malignant progression.
Our study has several limitations. First, data of this study were based on a population treated at a single quaternary academic medical center, which might have introduced institutional bias in algorithm building. Future endeavors are needed to validate this algorithm in a multi-institutional study to ensure that the algorithm is generalizable to the overall population. Second, variables included in the model were limited to demographic, clinical, and descriptive imaging characteristics. In future studies, genetic information, cyst aspirate characteristics, and quantitative imaging descriptors might need to be incorporated to potentially improve the performance of the model [
21]. In addition, this study did not stratify IPMNs based on the length of time from diagnosis to resection. Accounting for the length of time from diagnosis to resection might improve the accuracy of future models. Our model was also developed using data from IPMNs that were surgically resected without including IPMNs that were not resected. Moreover, this study was retrospective in nature which might have contributed to selection bias for patients who had a higher likelihood for surgical resection versus those with less worrisome features who could be followed by imaging surveillance alone.
In conclusion, IPMN-LEARN is a linear SVM-based machine learning model that might be useful as a complement to existing clinical guidelines to better determine which patients diagnosed with an IPMN might benefit from observation vs. surgical resection. It appeared to show high specificity, high sensitivity, and high positive predictive value when an independent set of patients were used to validate the model. Multi-institutional validation and testing are needed in the future.
Go to :






 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download