Abstract
Objective
To compare the degree of efficiency between density gradient centrifugation (DGC) method and an extended horizontal swim-up (SU) method.
Methods
A total of 97 couples undergoing in vitro fertilization were enrolled in the study. Semen samples were divided into three aliquots and treated using DGC, extended horizontal SU, and combined methods. DNA fragmentation and chromatin decondensation were detected in native semen samples and their three corresponding aliquots. The corresponding mature oocytes of each semen sample were divided into two sibling cultures. The first sibling culture was microinjected with semen pellets from DGC, and the second sibling culture was microinjected with semen pellets from the combination of both methods. Fertilization rate and embryonic development were assessed at day 3.
Results
DNA fragmentation and chromatin decondensation was significantly low in DGC and extended horizontal SU samples; however, the rates of DNA fragmentation and chromatin decondensation were significantly lower in extended horizontal SU samples than in DGC samples. The lowest rates of DNA fragmentation and chromatin decondensation corresponded to the samples treated with both methods. The highest rates of DNA fragmentation and chromatin decondensation corresponded to the samples treated with DGC. No significant difference was found in the fertilization rate or day 3 embryos between sibling cultures.
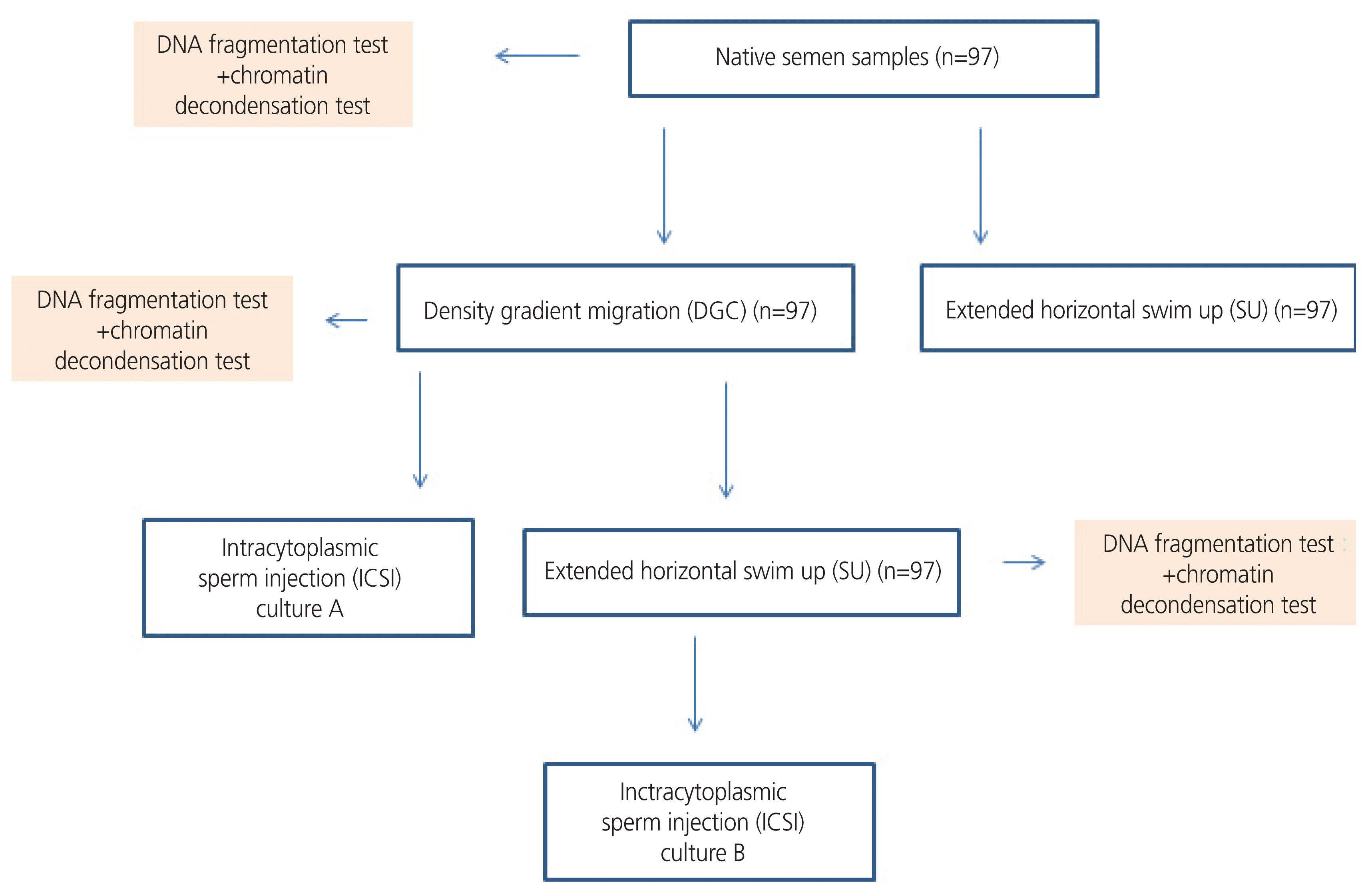
Sperm DNA integrity is considered the most important indicative parameter for distinguishing between infertile and fertile males and has better diagnostic and prognostic value than routine semen parameters [1,2]. Its assessment has recently been the subject of a large number of in-depth studies aimed at determining its impact on male infertility and in vitro fertilization (IVF) outcomes [3,4]. Semen DNA integrity is reflected in DNA fragmentation and chromatin decondensation. DNA fragmentation can occur at mitochondrial or nuclear levels. It can originate from apoptosis during spermatogenesis or the deleterious effects of reactive oxygen species (ROS) during sperm transport in the seminiferous tubules [5]. Furthermore, apoptotic nucleases and ROS can produce DNA breaks in ejaculated mature spermatozoa [6]. DNA fragmentation in ejaculated spermatozoa can also be explained by another mechanism, which is a defect in nuclear remodeling resulting in problems in the replacement of histones with protamines during spermiogenesis [6]. Chromatin decondensation mostly involves protamine deficiency and is characterized by chromatin compaction and DNA damage [7]. DNA fragmentation and abnormal chromatin condensation have been shown to affect sperm fertilizing ability and, consequently, embryo quality and implantation rates [8,9]. In assisted reproductive technology (ART), sperm preparation methods are being constantly developed to provide the best quality spermatozoa prior to oocyte fertilization. The most widely used sperm preparation techniques in ART laboratories are density gradient centrifugation (DGC) and swim-up (SU). These techniques, whether used separately or in combination, have been shown to improve sperm motility and morphology [10,11], as well as fertilization rates and embryonic quality [12,13]. However, regarding DNA integrity, the results available in the literature are contradictory. Some studies have shown that DGC can increase the production of ROS [14,15], and SU might be unable to provide sperm without causing DNA damage [16]. Other studies have shown that DGC and SU can select spermatozoa with the longest telomeres [17,18] and thereby decreasing the rate of DNA damage [19]. Therefore, this study aimed to elucidate the degree of efficiency of DGC in terms of DNA integrity and to compare these results with those of a novel extended horizontal SU technique (Fig. 1).
This study included 97 couples with primary/secondary infertility who underwent oocyte retrieval cycles of assisted reproduction between January 2021 and August 2021 (Table 1). Only normozoospermic semen samples in terms of numeration, mobility, and motility were included according to the World Health Organization 2010 criteria (numeration >20 M/mL, progressive motility ≥32%, and typical morphology ≥4%). All couples provided signed informed consent before the IVF cycle and sperm collection. Female patients with poor ovarian reserve were excluded from this study due to the difficulty of dividing those “low oocytes number” cultures into sibling cultures.
Women underwent personalized controlled ovarian stimulation using a gonadotropin-releasing hormone (GnRH) antagonist protocol. Initially, a daily subcutaneous injection of recombinant follicle-stimulating hormone (150–300 international unit [IU] recombinant follicle-stimulating hormone [rFSH]) (rFSH, Merck-Serono, Modugno, Italy) was used alone or in combination with human menopausal gonadotropin (Menopur, Ferring, Saint Prex, Switzerland). Then, the rFSH doses with/without human menopausal gonadotropin for ovarian hyperstimulation were calculated based on women’s age and anti-Müllerian hormone concentration, in addition to prior history of ovarian stimulation, and were adjusted according to usual parameters of follicle growth, determined by serum estradiol (E2) concentration and ultrasound monitoring.
A daily dose of GnRH antagonist (Cetrotide, Merck-Serono, Modugno, Italy or Orgalutran, MSD, Brussels, Belgium) was injected subcutaneously, starting from day 6 of FSH administration. The ovulation trigger was performed using 10,000 IU of human chorionic gonadotropin (Ovitrelle, Merck-Serono, Modugno, Italy) and gonadotropin-releasing hormone (Decapeptyl, Ferring, Saint Prex, Switzerland) after obtaining follicles that reached dimensions of 17 mm or greater in diameter and adequate serum E2 levels. Oocytes were retrieved 34–36 hours after human chorionic gonadotropin administration.
The retrieved oocytes were isolated from the follicular fluid, rinsed, and cultured in media (SAGE 1-Step, Origio, Malov, Denmark). Then, 2 to 3 hours after retrieval, the oocyte-corona-cumulus complexes were placed in a 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid-buffered medium (Ferticult Flushing medium, Fertipro, Beernem, Belgium) containing hyaluronidase (Hyaluronidases in Ferticult Flushing medium, Fertipro, Beernem, Belgium) and were mechanically denudated using a 20–200 μL micropipette. Only oocytes in the metaphase II stage underwent intracytoplasmic sperm injection (ICSI).
Sperm samples were collected from the male partner by masturbation in a sterile container after 3–4 days of abstinence. Semen emission from each patient was performed twice to collect sufficient samples for the study. After 30 minutes of liquefaction at room temperature, semen analysis was performed to assess numeration, motility, and morphology. Sperm count (M/mL) and motility (%) were performed from 20 μL of each sample in a 20 μm Makler counting chamber (Sefi Medical Instruments, Haifa, Israel). Semen motility was then observed under a 40xmagnification of a phase contrast microscope coupled with computer assisted sperm analysis software. For sperm morphology assessment, smears of raw samples were stained using a Diff-QUIK kit (Baxter Healthcare Corporation. Inc., McGaw, IL, USA) and rinsed in distilled water. The morphology was evaluated under 1,000xmagnification.
Aliquots from each ejaculate were collected for DGC, extended horizontal SU, and sperm DNA integrity assay.
The bottom fraction of the Puresperm (Nidacon International, Mölndal, Sweden) gradient (70%) was prepared by mixing 7 mL of commercially supplied Puresperm solution with 3 mL Earl’s balanced salt solution. The upper fraction (40%) was prepared by mixing 4 mL Puresperm with 6 mL Earl’s balanced salt solution. Briefly, 2 mL of liquefied semen was loaded onto 40% and 70% discontinuous gradients (each 1 mL) and centrifuged at 500 rpm for 20 minutes at room temperature. The pellet was resuspended in a fertilization medium (Sequential Fert™, Origio®, Malov, Denmark).
In a Petri dish, a zigzag trajectory (Fig. 2) was drawn with a buffered medium (Ferticult Heppes, FERTIPRO, Beernem, Belgium) using a 200 IU micropipette. The preparation was covered with mineral oil and heated at 37°C. Semen samples (40 μL), either from unprocessed semen or pellets of density gradient migration, were placed in the first drop of the designed horizontal SU (Fig. 2). Then the migration dish was incubated at 37°C for 30 minutes.
DNA fragmentation and chromatin decondensation were detected in the spare suspensions used for ART procedures (Fig. 1).
First, 50 μL of sperm were centrifuged with 200 μL of diluted phosphate-buffered saline (PBS) (1 mL of PBS for 8 mL of distilled water) at 1,200 rpm for 10 minutes. Then, each pellet was spread on two slides (DNA fragmentation analysis/chromatin decondensation analysis) and dried on a hotplate. The slides were then fixed with 1% formaldehyde for 30 minutes at 37°C and rinsed with water. The spermatozoa were permeabilized with an “enzyme” solution (100 μL of tritonx100+100 μL of crystallizing citrate+9,800 μL of distilled water) and then rinsed with water.
First, 5 μL of hydration solution (In Situ Cell Death Detection Kit, Fluorescein, Darmstadt, Germany) was placed on the slide and incubated for 45 minutes at 37°C in the dark. The slides were then rinsed with water. A drop of glycerol was added to the slide and incubated at 4°C for 1 hour. The readings were performed underx100 magnification of a fluorescence microscope (Nikon Eclipse 80i, Tokyo, Japan). At least 200 sperm cells from each sample were counted, and the percentage of TdT-mediated-dUTP nick-end labeling-positive cells was calculated.
First, 1 mL of aniline blue was deposited on the slides for 18 minutes at room temperature. The slides were then rinsed with tap water and dried. The reading was performed underx100 magnification of a white light microscope. The percentage of dark blue-colored sperm was determined relative to colorless (or weakly colored) sperm.
Mature oocytes from each patient were divided into two sibling cultures: culture A corresponded to oocytes microinjected with semen pellets from DGC, and culture B corresponded to oocytes microinjected with semen samples from the combination of DGC and extended horizontal SU. No oocytes were fertilized with spermatozoa from unprocessed semen.
The fertilization rate was calculated as the number of 2 pronuclei embryos on day 1 divided by the total number of metaphase II oocytes. The rate of 8 cells embryos on day 3 was calculated by dividing the number of 8 cells embryos by the total number of embryos.
The temperature inside the incubators (IVF-Cube AD3100, ASTEC, Fukuoka, Japan; HeraCell 150, Thermo Fisher scientific, Waltham, MA, USA) was controlled using a certified thermometer and maintained at 37±0.2°C. The oxygen level inside the incubators was 5%, and the cultivating medium pH was 7.3±0.02, with carbon dioxide at approximately 5.6%.
We noticed that the rates of DNA fragmentation and chromatin decondensation significantly decreased after DGC (P=6.77×10−19; P=1.42×10−14) and extended horizontal SU (P=1.63×10−29; P=2.09×10−28) (Table 2).
Comparing the two methods, the rates of DNA fragmentation and chromatin decondensation were significantly lower after extended horizontal SU than after DGC (P=1.29×10−33; P=2.23×10−25) (Table 3).
Comparing the results obtained after DGC alone, extended horizontal SU alone, and the combination of the two methods, we noticed that the lowest rates of DNA fragmentation and chromatin decondensation corresponded to the combination samples. On the other hand, the highest rate of DNA fragmentation and chromatin decondensation corresponded to the DGC samples (P=1.0×10−33; P=2.23×10−25) (Table 3).
We found no significant difference in the fertilization rate and day 3 embryo rate between the cultures corresponding to DGC semen samples (culture A) and the combination of DGC and extended horizontal SU samples (culture B) (Table 4).
DNA integrity can be altered by many factors, including errors in spermiogenesis, poor chromatin compaction, apoptosis, oxidative stress, and external factors, such as lifestyle, infection, and radiation [20,21]. Several studies have shown that increased DNA fragmentation and chromatin condensation in sperm has deleterious effects on IVF outcomes, from fertilization to pregnancy [3,11,22]. DGC and standard SU are the most common sperm treatment techniques routinely used in IVF laboratories worldwide [19,23]. It has been well established that these washing techniques ameliorate sperm quality. However, the available results in the literature are contradictory. When compared to conventional SU, the DGC technique has been shown to provide better capacitation and better responsiveness to calcium induction of the acrosome reaction [23] and to lower the rate of DNA damage [24,25]. Furthermore, other studies have shown that DGC increases ROS production [15,19,26,27] through the sheering forces generated by centrifugation [28]. Thus, ROS production resulting from lipid peroxidation within sperm membranes leads to the disruption of DNA integrity [19]. On the other hand, conventional SU has been shown to select spermatozoa with the longest telomeres, which is an indicator of correct spermatogenesis [18], decreasing the rate of apoptotic and necrotic cells in sperm [4]. However, it has been shown that the conventional SU method can also lead to ROS production due to increased cell-to-cell contact within spermatozoa [29]. According to previous research, none of these techniques have been unequivocally proven to avoid DNA damage. Instead, it has been proven to have the ability to decrease DNA damage but also produces de novo ROS under in vitro conditions. The conflicting evidence might give the impression of a vicious cycle that made us wonder what is more impactful between sperm selection by treatment techniques or the production of DNA damage.
There are several variants of horizontal sperm migration, such as the typical single straight medium line used in ICSI plates [30,31], side migration technique [32], and the one developed by Baldini et al. [33], using three medium drops connected through a medium bridge. Our extended horizontal SU involved a long journey for sperm, as spermatozoa had to go through a long line and make three turns before arriving at the endpoint. Thereforem the aim of our new approach was to elongate the path of spermatozoa migration to optimize sperm quality prior to DNA analysis.
Our data highlight the degree of attenuation of the rate of DNA deletions by DGC and extended horizontal SU. Our study showed that the rate of DNA fragmentation and chromatin decondensation significantly decreased after DGC treatment alone and extended horizontal SU treatment alone. However, while comparing DGC and extended horizontal SU, the last one was the sperm treatment that resulted in significantly the lowest rate of DNA deletions in terms of DNA fragmentations and chromatin decondensation. According to these initial results, DGC and extended horizontal SU were both capable of deselecting DNA damage and ameliorating DNA quality. However, we suggest that the extended horizontal SU alone is sufficient for normospermic patient selection before ICSI, mostly because they already have a low percentage of DNA deletions. Furthermore, since there was still a considerable percentage of DNA deletions after these two treatment methods, we combined both treatment methods to evaluate any differences. Our data showed that when both methods were combined, the decrease in the rates of DNA fragmentation and chromatin decondensation was significantly more important than after each method alone. Supporting these results, previous studies have shown that combining the two washing methods can lower the percentage of DNA fragmentation [34,35]. However, our study used a different extended horizontal SU than a well-adopted vertical SU.
Regarding IVF outcomes, we noted no correlation between embryo quality and the rate of DNA fragmentation or chromatin decondensation [27]. Because ICSI procedures were assessed after DGC for both sibling embryo cultures (cultures A and B), semen samples already had a lower rate of DNA fragmentation and chromatin decondensation. In addition to the ICSI procedure, sperm selection was performed according to morphological and motility features. Some studies have shown that the morphology of spermatozoa can be associated with DNA damage, as spermatozoa with normal morphology are less prone to DNA damage [36,37]. Based on these results, we can speculate that the DGC technique may be sufficiently efficient before ICSI because there is morphology-and motility-based selection that lowers the rate of DNA damage. Furthermore, other studies had shown no significant difference between the decreased rate of DNA fragmentation and poor IVF outcomes when the ICSI procedure was used, while differences were noted after conventional IVF [12,38]. However, the difference in embryo quality between the conventional IVF and ICSI procedures was not investigated in our study.
In conclusion, our study suggests that the combination of DGC and extended horizontal SU techniques significantly lowered the rates of DNA fragmentation and chromatin decondensation. If we had to choose between DGC alone or in combination with both methods, we would suggest that DGC alone might be sufficient for healthy embryo development. Again, we are aware that the current study had some limitations. First, the cultures were not extended to day five to investigate any association with the blastulation rate. Another limitation is the implantation rate data, as many studies have previously shown that the consequences of DNA damage are more prone to appear in the rates of implantation than in embryo morphology [39,40]. More studies are needed to confirm whether the addition of extended horizontal SU to sperm treatment is associated with significant positive IVF outcomes. However, based on our favorable data, we can still apply this method to more patients to improve sperm DNA quality before ICSI.
References
1. Evgeni E, Lymberopoulos G, Touloupidis S, Asimakopoulos B. Sperm nuclear DNA fragmentation and its association with semen quality in Greek men. Andrologia. 2015; 47:1166–74.

2. Montjean D, Zini A, Ravel C, Belloc S, Dalleac A, Copin H, et al. Sperm global DNA methylation level: association with semen parameters and genome integrity. Andrology. 2015; 3:235–40.

3. Oseguera-López I, Ruiz-Díaz S, Ramos-Ibeas P, Pérez-Cerezales S. Novel techniques of sperm selection for improving IVF and ICSI outcomes. Front Cell Dev Biol. 2019; 7:298.

4. Saylan A, Erimsah S. High quality human sperm selection for IVF: a study on sperm chromatin condensation. Acta Histochem. 2019; 121:798–803.

5. Li MW, Lloyd KCK. DNA fragmentation index (DFI) as a measure of sperm quality and fertility in mice. Sci Rep. 2020; 10:3833.

6. Muratori M, Marchiani S, Tamburrino L, Baldi E. Sperm DNA fragmentation: mechanisms of origin. Adv Exp Med Biol. 2019; 1166:75–85.

7. Dutta S, Henkel R, Agarwal A. Comparative analysis of tests used to assess sperm chromatin integrity and DNA fragmentation. Andrologia. 2021; 53:e13718.

8. Osman A, Alsomait H, Seshadri S, El-Toukhy T, Khalaf Y. The effect of sperm DNA fragmentation on live birth rate after IVF or ICSI: a systematic review and meta-analysis. Reprod Biomed Online. 2015; 30:120–7.

9. Galotto C, Cambiasso MY, Julianelli VL, Valzacchi GJR, Rolando RN, Rodriguez ML, et al. Human sperm decondensation in vitro is related to cleavage rate and embryo quality in IVF. J Assist Reprod Genet. 2019; 36:2345–55.

10. Fácio CL, Previato LF, Machado-Paula LA, Matheus PC, Filho Araújo E. Comparison of two sperm processing techniques for low complexity assisted fertilization: sperm washing followed by swim-up and discontinuous density gradient centrifugation. JBRA Assist Reprod. 2016; 20:206–11.
11. Oleszczuk K, Giwercman A, Bungum M. Sperm chromatin structure assay in prediction of in vitro fertilization outcome. Andrology. 2016; 4:290–6.

12. Borini A, Tarozzi N, Bizzaro D, Bonu MA, Fava L, Flamigni C, et al. Sperm DNA fragmentation: paternal effect on early post-implantation embryo development in ART. Hum Reprod. 2006; 21:2876–81.

13. Kim EK, Kim EH, Kim EA, Lee KA, Shin JE, Kwon H. Comparison of the effect of different media on the clinical outcomes of the density-gradient centrifugation/swim-up and swim-up methods. Clin Exp Reprod Med. 2015; 42:22–9.

14. Aitken RJ, Finnie JM, Muscio L, Whiting S, Connaughton HS, Kuczera L, et al. Potential importance of transition metals in the induction of DNA damage by sperm preparation media. Hum Reprod. 2014; 29:2136–47.

15. Álvarez-Rodríguez M, Álvarez M, Anel-López L, López-Urueña E, Manrique P, Borragán S, et al. Effect of colloid (androcoll-bear, percoll, and puresperm) selection on the freezability of brown bear (ursus arctos) sperm. Theriogenology. 2016; 85:1097–105.

16. Oguz Y, Guler I, Erdem A, Mutlu MF, Gumuslu S, Oktem M, et al. The effect of swim-up and gradient sperm preparation techniques on deoxyribonucleic acid (DNA) fragmentation in subfertile patients. J Assist Reprod Genet. 2018; 35:1083–9.

17. Zhao F, Yang Q, Shi S, Luo X, Sun Y. Semen preparation methods and sperm telomere length: density gradient centrifugation versus the swim up procedure. Sci Rep. 2016; 6:39051.

18. Rocca MS, Speltra E, Menegazzo M, Garolla A, Foresta C, Ferlin A. Sperm telomere length as a parameter of sperm quality in normozoospermic men. Hum Reprod. 2016; 31:1158–63.

19. Lestari SW, Lestari SH, Pujianto DA. Sperm quality after swim up and density gradient centrifugation sperm preparation with supplementation of alpha lipoic acid (ALA): a preliminary study. AIP Conf Proc. 2018; 1933:030015.

20. Aitken RJ, Bronson R, Smith TB, De Iuliis GN. The source and significance of dna damage in human spermatozoa; a commentary on diagnostic strategies and straw man fallacies. Mol Hum Reprod. 2013; 19:475–85.

21. Agarwal A, Roychoudhury S, Bjugstad KB, Cho CL. Oxidation-reduction potential of semen: what is its role in the treatment of male infertility? Ther Adv Urol. 2016; 8:302–18.

22. Simon L, Zini A, Dyachenko A, Ciampi A, Carrell DT. A systematic review and meta-analysis to determine the effect of sperm DNA damage on in vitro fertilization and intracytoplasmic sperm injection outcome. Asian J Androl. 2017; 19:80–90.

23. Hernández-Silva G, López-Torres AS, Maldonado-Rosas I, Mata-Martínez E, Larrea F, Torres-Flores V, et al. Effects of semen processing on sperm function: differences between swim-up and density gradient centrifugation. World J Mens Health. 2021; 39:740–9.

24. Parmegiani L, Cognigni GE, Filicori M. Sperm selection: effect on sperm DNA quality. Adv Exp Med Biol. 2014; 791:151–72.

25. Xue X, Wang WS, Shi JZ, Zhang SL, Zhao WQ, Shi WH, et al. Efficacy of swim-up versus density gradient centrifugation in improving sperm deformity rate and DNA fragmentation index in semen samples from teratozoospermic patients. J Assist Reprod Genet. 2014; 31:1161–6.

26. Li J, Ning B, Cao X, Luo Y, Guo L, Wei G, et al. Separation of motile sperm for in vitro fertilization from frozen-thawed bull semen using progesterone induction on a microchip. Anim Reprod Sci. 2016; 172:52–9.

27. Palini S, Stefani S, Primiterra M, Benedetti S, Barone S, Carli L, et al. Comparison of in vitro fertilization outcomes in ICSI cycles after human sperm preparation by density gradient centrifugation and direct micro swim-up without centrifugation. JBRA Assist Reprod. 2017; 21:89–93.
28. Aitken RJ, De Iuliis GN, Finnie JM, Hedges A, McLachlan RI. Analysis of the relationships between oxidative stress, DNA damage and sperm vitality in a patient population: development of diagnostic criteria. Hum Reprod. 2010; 25:2415–26.

29. Takeshima T, Yumura Y, Kuroda S, Kawahara T, Uemura H, Iwasaki A. Effect of density gradient centrifugation on reactive oxygen species in human semen. Syst Biol Reprod Med. 2017; 63:192–8.

30. Fréour T, Barragan M, Ferrer-Vaquer A, Rodríguez A, Vassena R. WBP2NL/PAWP mRNA and protein expression in sperm cells are not related to semen parameters, fertilization rate, or reproductive outcome. J Assist Reprod Genet. 2017; 34:803–10.

31. Baldini D, Ferri D, Baldini GM, Lot D, Catino A, Vizziello D, et al. Sperm selection for icsi: do we have a winner? Cells. 2021; 10:3566.

32. Hinting A, Lunardhi H. Better sperm selection for intracytoplasmic sperm injection with the side migration technique. Andrologia. 2001; 33:343–6.

33. Baldini D, Baldini A, Silvestris E, Vizziello G, Ferri D, Vizziello D. A fast and safe technique for sperm preparation in ICSI treatments within a randomized controlled trial (RCT). Reprod Biol Endocrinol. 2020; 18:88.

34. Berteli TS, Da Broi MG, Martins WP, Ferriani RA, Navarro PA. Magnetic-activated cell sorting before density gradient centrifugation improves recovery of high-quality spermatozoa. Andrology. 2017; 5:776–82.

35. Esbert M, Godo A, Soares SR, Florensa M, Amorós D, Ballesteros A, et al. Spermatozoa with numerical chromosomal abnormalities are more prone to be retained by annexin V-MACS columns. Andrology. 2017; 5:807–13.

36. Moskovtsev SI, Willis J, White J, Mullen JB. Sperm DNA damage: correlation to severity of semen abnormalities. Urology. 2009; 74:789–93.

37. Enciso M, Iglesias M, Galán I, Sarasa J, Gosálvez A, Gosálvez J. The ability of sperm selection techniques to remove single- or double-strand DNA damage. Asian J Androl. 2011; 13:764–8.

38. Pregl Breznik B, Kovačič B, Vlaisavljevič V. Are sperm DNA fragmentation, hyperactivation, and hyaluronan-binding ability predictive for fertilization and embryo development in in vitro fertilization and intracytoplasmic sperm injection? Fertil Steril. 2013; 99:1233–41.
39. Borges E Jr, Zanetti BF, Setti AS, Braga DPAF, Provenza RR, Iaconelli A Jr. Sperm DNA fragmentation is correlated with poor embryo development, lower implantation rate, and higher miscarriage rate in reproductive cycles of non-male factor infertility. Fertil Steril. 2019; 112:483–90.

40. Casanovas A, Ribas-Maynou J, Lara-Cerrillo S, Jimenez-Macedo AR, Hortal O, Benet J, et al. Double-stranded sperm DNA damage is a cause of delay in embryo development and can impair implantation rates. Fertil Steril. 2019; 111:699–707e1.
Fig. 2
Representation of the extended swim up by culture medium in a petri plate. (A) First drop where the sperm is added. (B) Second drop where sperm is migrating toward.
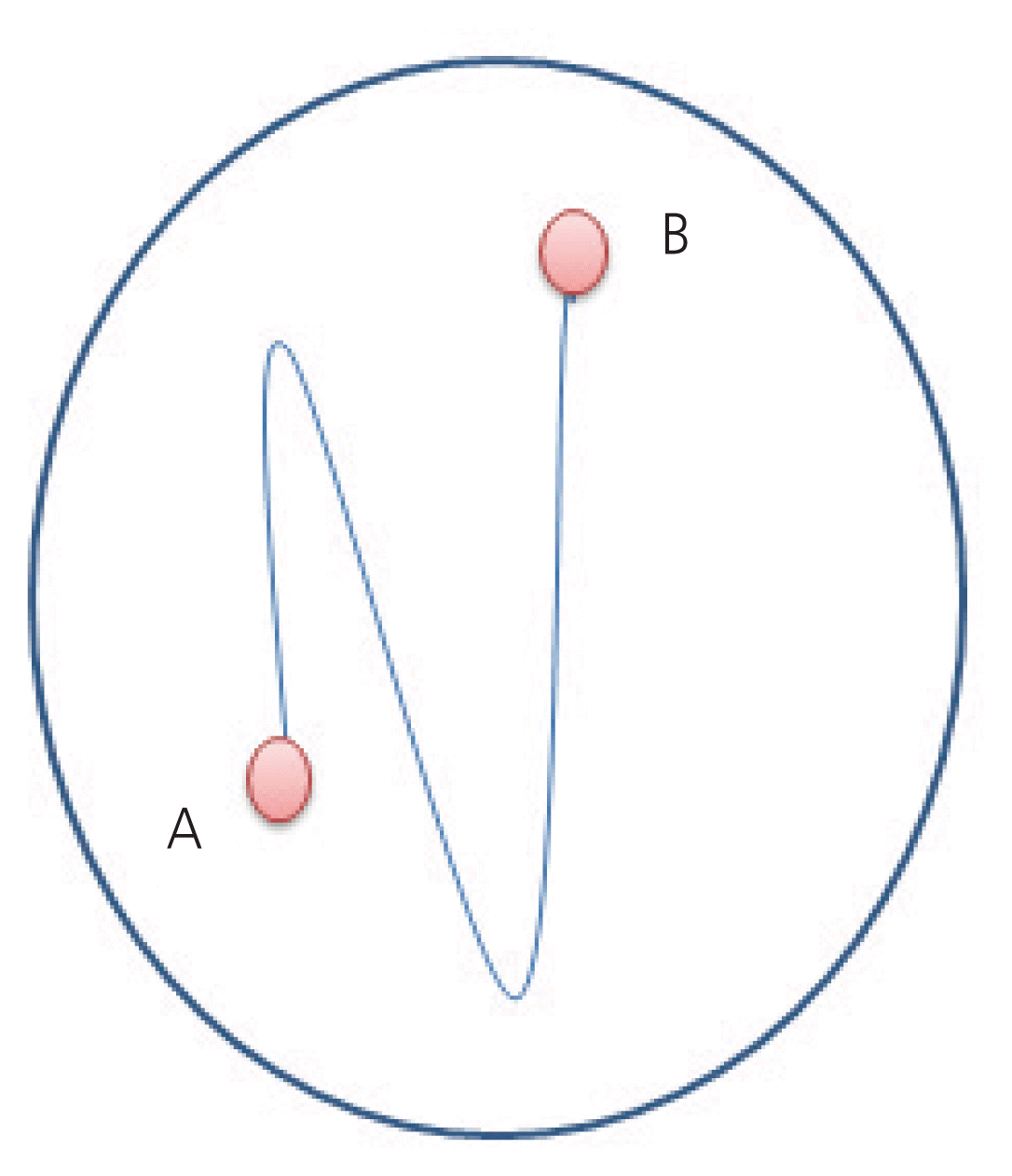
Table 1
Demographic characteristics of patients (97)
Table 2
Comparison of the rate of DNA fragmentation/chromatin decondensation before and after density gradient migration, extended horizontal swim up, and the combination of both methods
Table 3
Comparison of the rate of DNA fragmentation and chromatin decondensation between density gradient migration, extended swim up, and the combination of both methods
Table 4
Comparison of the rate of fertilization and day 3 embryos between culture A (ISCI with semen from density gradient migration) and culture B (ICSI with semen from the combination of density gradient migration and extended swim-up method)
| Culture A | Culture B | P | |
|---|---|---|---|
| Fertilization rate (n=61) | 74.11±27.91 | 71.72±31.05 | NS |
| Rate of 8 cells embryos on day 3 (n=61) | 68.76±32.44 | 60.03±36.74 | NS |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download