1. Model Performance
As can be observed from
Table 2, our method outperformed the previous state-of-the-art results in both traditional accuracy and balanced accuracy, except for the results reported by Jaworek-Korjakowska et al. [
11]. We will scrutinize those results in detail in the next section. The highest balanced accuracy was attained when the method was trained on four classes and used to predict the partially merged three classes. The problem of categorizing into four categories is a more complex problem, which is evident from the numerical values.
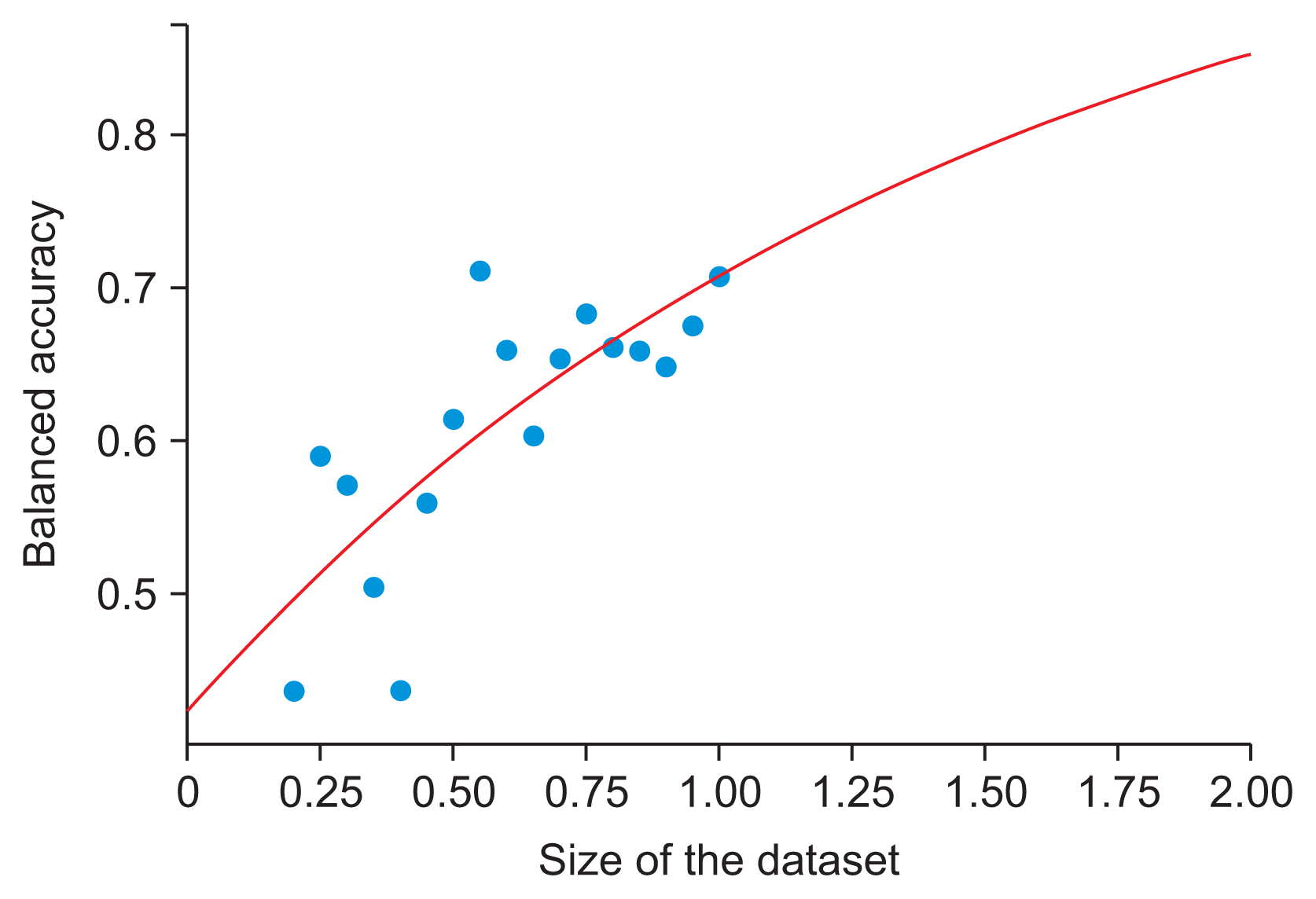
Although it is commonly believed that stronger backbones perform better, this is not always the case, as can be seen in
Table 3. Multiple factors might explain why the balanced accuracy obtained with EfficientNet-B5 was lower than that of B4: (1) the larger backbone (B5) contained more parameters, making it more liable to overtraining, especially for the small dataset we used; (2) the uncertainty in the performance values is large for a small dataset, even if cross-validation is used (for balanced accuracy, the error contribution of the minority classes can be especially large); and (3) our hyper-parameters were optimized more heavily for B4. We note that uncertainties can, in principle, be evaluated by conducting a series of independent training sessions with varying random dataset splits for cross-validation, and then comparing the outcomes [
16].
2. Data Leakage in [11] and its Reproduction
This section presents our analysis demonstrating that the high scores reported by Jaworek-Korjakowska et al. [
11] were most probably due to data leakage.
Jaworek-Korjakowska et al. [
11] offered a thorough account of the pipeline they used for both training and validating purposes in their paper. Following image processing such as cropping and resizing, each image was allocated to one of three thickness classes. These categories were identical to ours, and our datasets shared similarities (
Table 1).
The authors used the SMOTE oversampling technique [
12] to deal with the class imbalance in the preprocessed data. They generated new samples for minority classes based on existing data in the given class, resulting in equal-size classes. Once the classes were balanced, the data were split into two parts: training data (to train the model) and testing data (for performance evaluation).
In machine learning, it is common practice to use separate data for training and for validation [
15]. This eliminates the possibility of “memorizing” the dataset, allowing an accurate estimation of model performance on new, unseen data. However, the training and validation datasets must be entirely independent for this to work. Data leakage arises when information about the validation data is incorporated into the training procedure, providing an unrealistic and overly optimistic estimate of the model’s performance. This leakage usually occurs when pre-processing the training and testing data jointly [
17].
The above problem is an issue for that paper, as the authors utilized SMOTE oversampling prior to dividing the data. SMOTE functions by generating synthetic data from a few representative samples from the same class to increase the minority classes to the size of the most prominent class. Although it is an appropriate method for low-dimensional data, SMOTE can create nearly identical copies of specific pre-existing images with visual data since the dimensionality of the problem space tends to be extremely high.
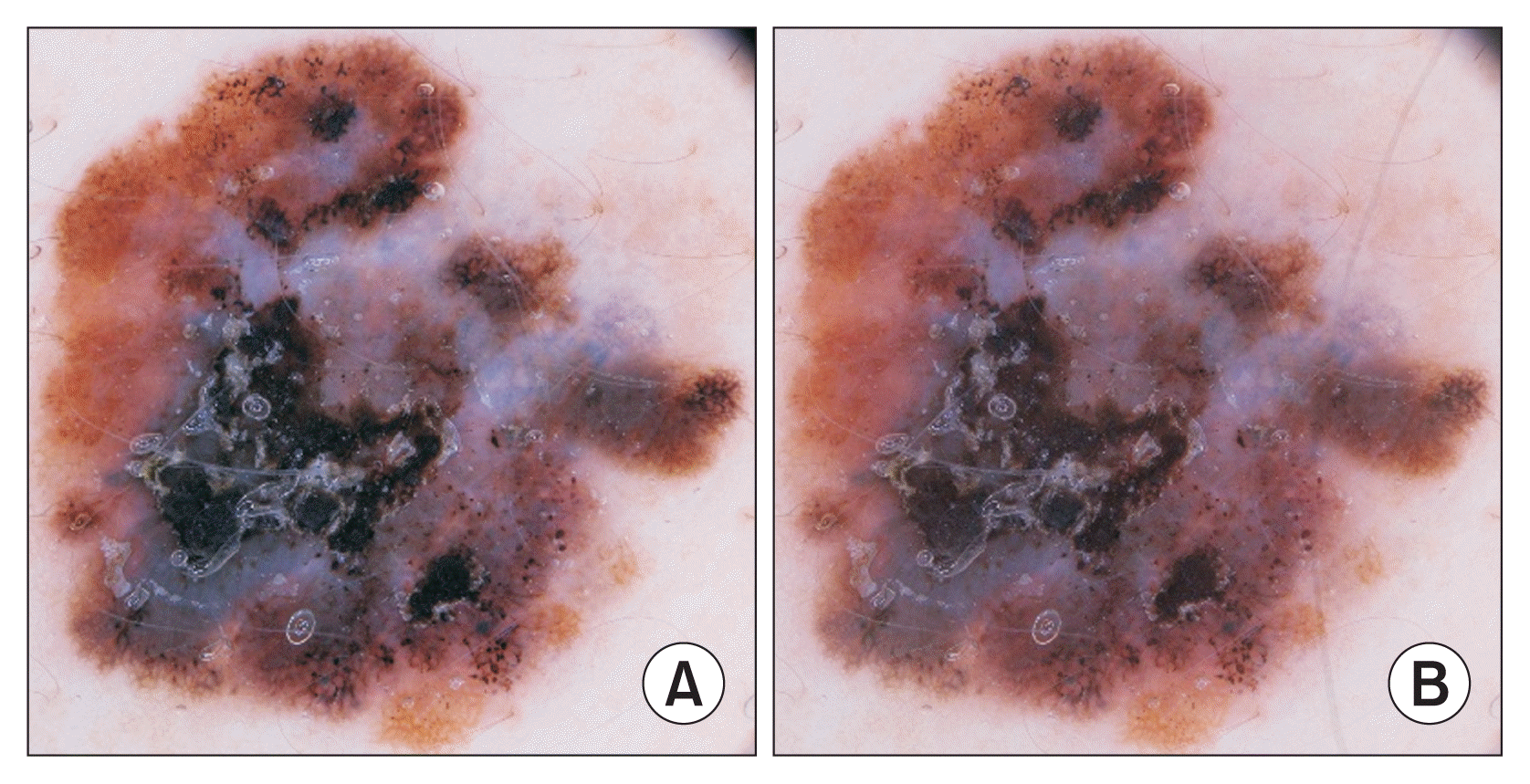
To illustrate, we applied the SMOTE algorithm to a collection of melanoma images. The minority class in the dataset comprised 28 samples, which we augmented by adding 74 artificially generated images, resulting in a total of 102 images. These numbers correspond to the size of the smallest minority class and the majority class in the dataset that we employed for our tests, respectively.
Figure 2 depicts two images: one of them belonged to the original dataset, whereas the other was produced by the SMOTE algorithm. Notably, the newly created image bears significant similarity to the original one. It can clearly be seen that by splitting the dataset after the oversampling step, the training set could include samples that have been influenced by some validation samples, resulting in a misleadingly high score during evaluation.
To prove our hypothesis regarding data leakage, we implemented the algorithm while carefully following every step. We then modified the algorithm slightly by only applying the SMOTE oversampling to the training set, while making sure not to mix the training and validation samples. We used five-fold cross-validation to obtain a more stable evaluation of the model’s actual performance.
In this experiment, we utilized the Derm7pt dataset [
13], which bears resemblances to the dataset presented by Jaworek-Korjakowska et al. [
11] (
Table 1). To enable three-class classification [
11], melanoma in situ and melanoma depth of 0.76 mm were merged.
Table 4 shows the performance of the method presented in this paper and the result we obtained using this method with or without data leakage (i.e., where the oversampling step was performed before the dataset splitting). Jaworek-Korjakowska et al. [
11] provided accuracy, the ATPR, the ATNR, and the F1-score as metrics to measure performance; thus, we compared these scores in our experiment.
Our results with data leakage were similar to those reported in the original paper and, in some cases, even surpassed them. However, when we tested the method without data leakage by only validating on images the model did not see during training, we obtained much lower values. This was particularly evident in the ATPR and F1-score, which, unlike accuracy, give equal weight to each class and provide a more meaningful metric for imbalanced datasets.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download