Dear Editor,
Kocuria spp. are gram-positive cocci formerly classified as members of Micrococcus spp. that belong to the family Micrococcaceae [1]. They are commonly found in the environment, including water and soil, and also exist in the mucosa and skin of animals [1–3]. Among Kocuria spp., Kocuria kristinae and Kocuria varians cause most human infections [2, 3]. Two cases of Kocuria rhizophila infection involving the central venous catheter (CVC) have been reported [4, 5]; however, studies reporting these cases analyzed less than 500 bp of the 16S ribosomal (r) RNA gene, limiting their accuracy. We report the first Korean case of catheter-related bloodstream infection (CRBSI) caused by K. rhizophila, confirmed by genomic evidence. The Institutional Review Board of Seoul National University Hospital, Seoul, Korea, approved this study (2205-142-1327) and waived the need for informed consent.
In January 2022, an 18-year-old boy with a vascular-access port for home parenteral nutrition (HPN) was hospitalized because of fever. He had been on HPN for four years because of an avoidant restrictive food intake disorder and was hospitalized for recurrent CRBSIs. On day 1, his temperature was 39.4°C, and a complete blood count indicated a leukocyte count of 3.58×109/L (reference interval [RI]: 4.0–10.0×109/L), with 84.9% neutrophils; the C-reactive protein level was 26.1 mg/L (RI: <5 mg/L). Two pairs of blood samples collected from the peripheral vein (PV) and one pair collected from the CVC were inoculated into BACTEC Peds Plus/F and Lytic/10 Anaerobic/F bottles (Becton Dickinson, Sparks, MD, USA) and incubated in a BACTEC FX Blood Culture System (Becton Dickinson). He received intravenous vancomycin 690 mg at six hours interval as empirical antibiotic therapy.
Gram-positive cocci were detected in all aerobic culture bottles. The times to positive results were 17 hours (CVC) and 73 and 75 hours (PV), suggesting CRBSI. Colonies on blood agar were smooth, circular, and cream-colored. The isolates were identified as K. rhizophila using the VITEK 2 GP ID card (bioMérieux, Durham, NC, USA), with an excellent accuracy (99.0%). It was identified at the genus level as K. rhizophila (score value: 1.936) by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) using MALDI Biotyper (Bruker Daltonics, Bremen, Germany).
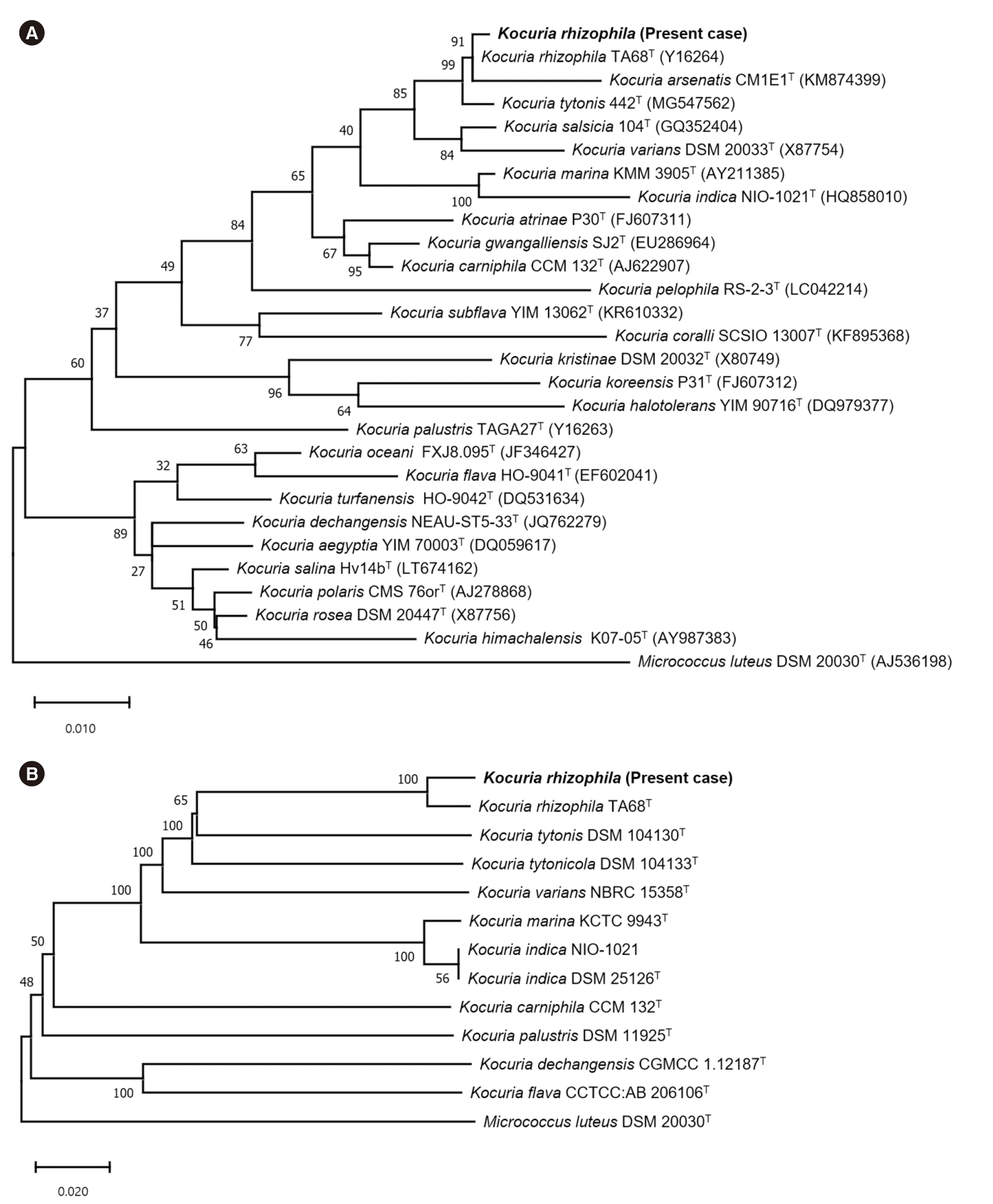
Next, 16S rRNA sequence analysis was performed, and the sequence was searched in the GenBank database and interpreted according to CLSI guidelines [6]. The sequence showed 99.93% identity with K. rhizophila (AY030315.1) and 99.79% identity with K. varians (MN072929.1). A phylogenetic tree of the 16S rRNA sequences of the isolate and type strains of the genus Kocuria constructed using MEGA-11 (http://www.megasoftware.net) showed that the isolate formed a clade with K. rhizophila (Fig. 1A). The results of antimicrobial susceptibility tests using Etest (bioMérieux) were interpreted as Micrococcus spp. or Staphylococcus spp. (Table 1) [7].
The fever subsided by the third day of hospitalization, and follow-up blood cultures on day 4 were negative. Because the chemo port was changed only three months prior, vancomycin-lock therapy was started on day 5 to preserve it. Intravenous vancomycin and vancomycin-lock therapy were administered until day 16. There were no additional symptoms after the discontinuation of vancomycin, and the patient was discharged on day 26.
Given the high similarity of 16S rRNA sequences of Kocuria spp., whole-genome sequencing (WGS) of the isolate using the MiSeq platform (Illumina, San Diego, CA, USA) was performed within the MAFGEN project [8]. The assembled genome size was 2,771,437 bp; the GC content was 70.57%. Analysis using TrueBac ID (CJ Bioscience Inc., Seoul, Korea) revealed that it shared the highest average nucleotide identity with K. rhizophila (97.76%), followed by K. tytonis (88.10%). The identification was confirmed by a genome-based phylogenetic tree built using the Type Strain Genome Server (TYGS) (Fig. 1B) [9]. The rpoB sequence (3,510 bp) was retrieved from GenBank; it showed 99.94% identity with K. rhizophila (CP072262.1), followed by 94.99% with K. varians (CP059343.1), indicating that the rpoB sequence is useful for identifying Kocuria [6].
Although Kocuria spp. are human commensals, the recent increase in cases of Kocuria infections indicates their pathogenic potential in immunocompromised or chronically catheterized patients [2–5]. Consistent with the previously reported K. rhizophila infections, the patient had a history of prolonged catheter use and recurrent CRBSIs [4, 5]. CRBSI is a common but significant complication in patients receiving HPN, especially in children [10].
While 16S rRNA sequencing is commonly used for bacterial identification, few organisms, including Kocuria spp., cannot be identified at the species level even with full-length 16S rRNA sequences [6]. WGS is increasingly used in clinical microbiology laboratories, and rare bacterial species can be accurately identified using TruBac ID or TYGS [8, 9]. This is the first case of CRBSI caused by K. rhizophila confirmed with genomic evidence in Korea.
The whole-genome sequence of the isolated bacterium was submitted to the BioSample database (accession No.: SAMN32639282).
Notes
AUTHOR CONTRIBUTIONS
Study conception and design: Yun KW and Park JH. Data acquisition: Kim Y. Data analysis and interpretation: Kim Y and Park JH. Figure preparation: Kim Y and Park JH. Manuscript drafting: Kim Y and Park JH. Manuscript revision: Kim Y, Kim TS, Park H, Yun KW, and Park JH. All authors read and approved the final manuscript.
REFERENCES
1. Li J, Zhang S. 2020; Kocuria coralli sp. nov., a novel actinobacterium isolated from coral reef seawater. Int J Syst Evol Microbiol. 70:785–9. DOI: 10.1099/ijsem.0.003825. PMID: 31671054.
2. Purty S, Saranathan R, Prashanth K, Narayanan K, Asir J, Sheela Devi C, et al. 2013; The expanding spectrum of human infections caused by Kocuria species: a case report and literature review. Emerg Microbes Infect. 2:e71. DOI: 10.1038/emi.2013.93. PMID: 26039557. PMCID: PMC3880876.
3. Kandi V, Palange P, Vaish R, Bhatti AB, Kale V, Kandi MR, et al. 2016; Emerging bacterial infection: identification and clinical significance of Kocuria species. Cureus. 8:e731. DOI: 10.7759/cureus.731.
4. Becker K, Rutsch F, Uekötter A, Kipp F, König J, Marquardt T, et al. 2008; Kocuria rhizophila adds to the emerging spectrum of micrococcal species involved in human infections. J Clin Microbiol. 46:3537–9. DOI: 10.1128/JCM.00823-08. PMID: 18614658. PMCID: PMC2566074.

5. Moissenet D, Becker K, Mérens A, Ferroni A, Dubern B, Vu-Thien H. 2012; Persistent bloodstream infection with Kocuria rhizophila related to a damaged central catheter. J Clin Microbiol. 50:1495–8. DOI: 10.1128/JCM.06038-11. PMID: 22259211. PMCID: PMC3318534.

6. CLSI. 2018. Interpretive criteria for identification of bacteria and fungi by targeted DNA sequencing. CLSI MM18. 2nd ed. Clinical and Laboratory Standards Institute;Wayne, PA:
7. CLSI. 2016. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. CLSI M45. 3rd ed. Clinical and Laboratory Standards Institute;Wayne, PA:
8. Ha SM, Kim CK, Roh J, Byun JH, Yang SJ, Choi SB, et al. 2019; Application of the whole genome-based bacterial identification system, TrueBac ID, using clinical isolates that were not identified with three matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems. Ann Lab Med. 39:530–6. DOI: 10.3343/alm.2019.39.6.530. PMID: 31240880. PMCID: PMC6660342.

9. Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M. 2022; TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 50:D801–7. DOI: 10.1093/nar/gkab902. PMID: 34634793. PMCID: PMC8728197.
10. Ross VM, Guenter P, Corrigan ML, Kovacevich D, Winkler MF, Resnick HE, et al. 2016; Central venous catheter infections in home parenteral nutrition patients: outcomes from Sustain: American Society for Parenteral and Enteral Nutrition's National Patient Registry for Nutrition Care. Am J Infect Control. 44:1462–8. DOI: 10.1016/j.ajic.2016.06.028. PMID: 27908433.

Fig. 1
Phylogenetic analysis of Kocuria isolates (type and reference strains) based on 16S rRNA gene (1,442 nucleotide positions) and genome sequences. (A) Phylogenetic tree constructed by the maximum-likelihood method using the model GTR+I+G and Micrococcus luteus DSM 20030T (AJ536198) as the outgroup. Bootstrap values are expressed as percentages of 1,000 replications, and the scale bar indicates the estimated number of substitutions per base. GenBank accession numbers are indicated in parentheses. (B) Phylogenetic tree constructed using the Type Strain Genome Server and M. luteus DSM 20030T as the outgroup. Branch lengths are scaled in terms of genome BLAST distance phylogeny method (GBDP) distance formula d5. Numbers above branches are GBDP pseudo-bootstrap support values from 100 replications




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download