Abstract
The fifth edition of the WHO classification (2022 WHO) and the International Consensus Classification (2022 ICC) of myeloid neoplasms have been recently published. We reviewed the changes in the diagnosis distribution in patients with MDS with excess blasts (MDS-EB) or AML using both classifications. Forty-seven patients previously diagnosed as having AML or MDS-EB with available mutation analysis data, including targeted next-generation and RNA-sequencing data, were included. We reclassified 15 (31.9%) and 27 (57.4%) patients based on the 2022 WHO and 2022 ICC, respectively. One patient was reclassified as having a translocation categorized as a rare recurring translocation in both classifications. Reclassification was mostly due to the addition of mutation-based diagnostic criteria (i.e., AML, myelodysplasia-related) or a new entity associated with TP53 mutation. In both classifications, MDS diagnosis required the confirmation of multi-hit TP53 alterations. Among 14 patients with TP53 mutations, 11 harbored multi-hit TP53 alterations, including four with TP53 mutations and loss of heterozygosity. Adverse prognosis was associated with multi-hit TP53 alterations (P=0.009) in patients with MDS-EB, emphasizing the importance of detecting the mutations at diagnosis. The implementation of these classifications may lead to the identification of different subtypes from previously heterogeneous diagnostic categories based on genetic characteristics.
Genome studies of myeloid hematological malignancies, including AML and MDS, have advanced our understanding of these diseases and revealed molecularly distinct groups [1–4]. Recently, the fifth edition of the WHO classification (2022 WHO) and the International Consensus Classification (2022 ICC) of myeloid neoplasms were published [5, 6]. In both classifications, the major changes regarding AML and MDS include: (1) changes in the blast thresholds defining AML, including the expansion of the genetic abnormality categories that may be diagnosed as AML with a blast cut-off requirement of 10%; (2) the introduction of an AML classification subtype termed “rare recurring translocation”; and (3) the extension of mutation-based definitions [5–7]. We reclassified patients diagnosed as having AML and MDS with excess blasts (MDS-EB) according to the 2016 WHO classification (2016 WHO), using the 2022 WHO and 2022 ICC. As the blast percentage defining MDS-EB and AML has been lowered and various genetic alterations have been introduced in both classifications, we reviewed the changes in the diagnosis distribution of patients with MDS-EB and AML using both the 2022 WHO and 2022 ICC. In addition, we focused on a newly introduced classification category, including TP53 mutation.
Forty-seven adult patients diagnosed as having AML or MDS-EB between November 2019 and September 2021 with available mutation analysis data, including targeted next-generation and RNA-sequencing data, were included in our study. Patients with AML with 2016 WHO-designated recurring balanced translocations were excluded (Fig. 1). This study was approved by the Institutional Review Board of Korea University, Guro Hospital, Seoul, Korea (2021GR0247, 2021GR0572). Overall patient characteristics are provided in Supplemental Data Table S1.
We retrospectively reviewed the clinical responses of all patients and conducted a survival analysis. Cytogenetic abnormalities were analyzed according to the 2020 International System for Human Cytogenomic Nomenclature guidelines [8]. Sequence data were mapped to the reference genome GrCH37 (hg19). Mutation analysis was performed using the Oncomine Myeloid Research Assay (Thermo Fisher Scientific, Waltham, MA, USA). Libraries were prepared using the Ion Chef System (Thermo Fisher Scientific) and sequenced using an Ion S5 XL sequencer (Thermo Fisher Scientific). Alignment and base calling were performed using Torrent Suite (v5.10), and variant calling was performed using Ion Reporter (v5.10). The variant allele fraction (VAF) cut-off for missense and nonsense variants was 5% and 10%, respectively, for insertions/deletions. RNA-sequencing libraries were prepared using the TruSeq Stranded Total RNA LT sample preparation kit (Illumina, San Diego, CA, USA) and sequenced using the NovaSeq platform (Illumina). Sequencing reads were aligned using the STAR aligner (v2.6.0), and fusion transcripts were detected using Arriba (v2.2.0) [9, 10]. For samples with TP53 mutations, a single-nucleotide polymorphism microarray analysis was performed using the CytoScan 750 K Assay (Affymetrix/Thermo Fisher). All statistical analyses were performed using R (v3.4.3; R Core Team, 2021) [11]. Detailed statistical methods are provided in the Supplemental Material.
Although the genetic abnormality categories that may be diagnosed as AML with a blast cut-off requirement of 10% have been expanded, patients with MDS-EB that were reclassified as AML were not observed. Among the 37 patients with AML, AML with recurrent genetic abnormalities accounted for 45.9% (17/37) according to the 2016 WHO, whereas 32.4% (12/37) of patients with AML were classified as AML with defining genetic abnormalities according to the 2022 WHO and as AML with recurrent genetic abnormalities according to the 2022 ICC (Fig. 2). Because of the exclusion of the provisional entity of AML with mutated RUNX1 from AML with recurrent genetic abnormalities in the 2022 WHO and 2022 ICC, the diagnoses of six patients with AML with RUNX1 mutations were modified. Notably, all of these six patients harbored mutations in myelodysplasia-related (MR) genes (e.g., SRSF2, BCOR, ASXL1, SF3B1, and U2AF1), and the diagnoses were reclassified to AML, MR according to the 2022 WHO, based on the mutation-based definition. In the 2022 ICC, RUNX1 is considered an MR gene; therefore, the diagnosis was reclassified as AML with MR gene mutations. One patient was reclassified as AML with FUS::ERG, as the translocation was categorized as a rare recurring translocation in both classifications. RNA-sequencing did not reveal other rare recurring translocations included in the 2022 WHO or 2022 ICC, probably because translocations are rare, and numerous translocations that occur predominantly in infants and children were included as rare recurring translocations [6, 8].
AML with myelodysplasia-related changes (AML-MRC) accounted for 24.3% (9/37) of the patients with AML according to the 2016 WHO; an increase in patients classified as AML, MR according to the 2022 WHO (45.9%, N=17/37) and AML with MR gene mutation or AML with MR cytogenetic abnormalities according to the 2022 ICC (32.4%, N=12/37) was noted when compared to the classification according to the 2016 WHO (Fig. 2). With the introduction of the mutation-based definition of AML, MR, in addition to patients with AML with mutated RUNX1, three patients with AML, not otherwise specified were reclassified as MR AML according to both the 2022 WHO and the 2022 ICC.
TP53 alterations, especially TP53 mutations, in MDS and AML are often associated with a complex karyotype and have highly adverse prognostic implications [3, 13–15]; therefore, the presence of TP53 mutations is recognized as a new category in both the 2022 WHO and the 2022 ICC. In the 2022 ICC, a disease category of myeloid neoplasms with mutated TP53 encompassing MDS, MDS/AML, and AML with mutated TP53 has been newly introduced as a distinct category. Regarding the 2022 WHO, patients with MDS with multi-hit TP53 alterations are classified as MDS with biallelic TP53 (MDS-biTP53), regardless of blast percentage; however, a distinct category for AML with mutated TP53 was not defined. Based on the 2022 ICC, five patients with AML-MRC, two patients with pure erythroid leukemia, and one patient with therapy-related myeloid neoplasms were reclassified as patients with AML with TP53 mutation, which accounted for 21.6% (8/37) of the patients with AML. Of the 10 patients with MDS-EB, five had multi-hit TP53 alterations and one had a single TP53 mutation. As multi-hit TP53 alteration status is a diagnostic criterion for MDS-biTP53, one patient with MDS-EB-2 with a single TP53 mutation was classified as MDS-IB-2 according to the 2022 WHO, whereas the classification according to the 2022 ICC was MDS/AML with mutated TP53.
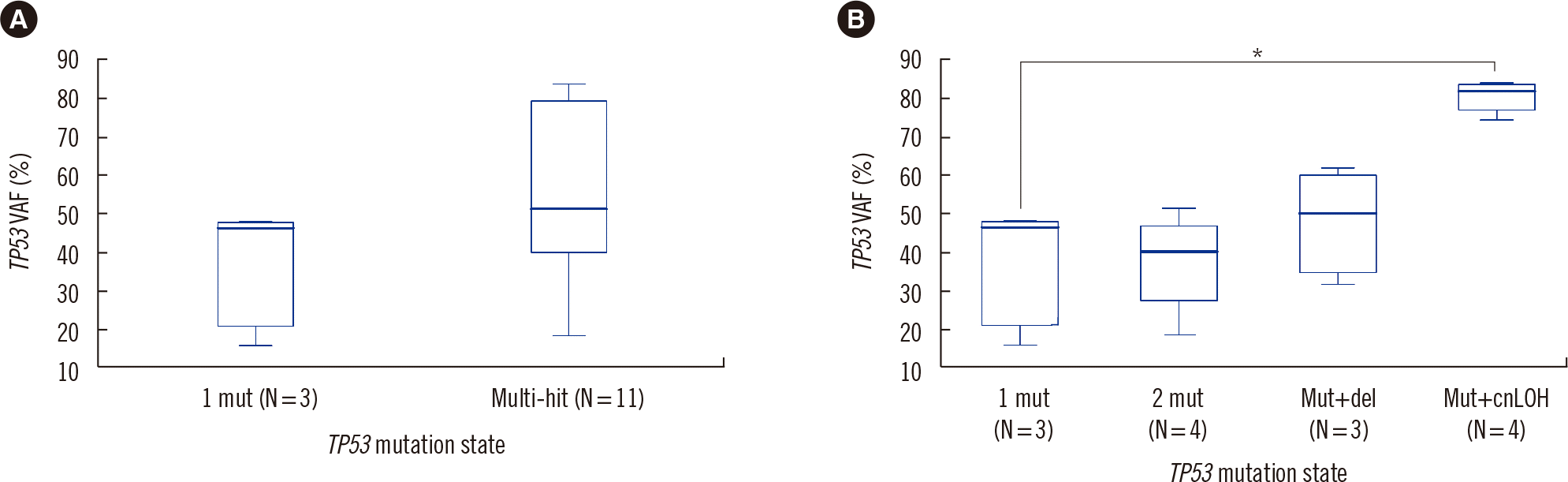
Among the 47 patients, 14 patients (six with MDS-EB, seven with AML, and one with therapy-related myeloid neoplasms) harbored TP53 mutations. All of these 14 patients had a complex karyotype (≥3 chromosomal abnormalities), and 11 patients harbored multi-hit TP53 alterations (three with two TP53 mutations, four with one TP53 mutation and copy number loss of TP53, and four with TP53 mutation and copy number neutral loss of heterozygosity [cnLOH]). TP53 VAF, estimated by the maximum TP53 VAF value for the multi-hit TP53 alteration group, did not significantly differ between the single TP53 mutation and multi-hit TP53 alteration groups; however, patients with multi-hit TP53 with cnLOH tended to have higher VAFs than those with a single TP53 mutation (P=0.057; median VAF, 46.5% vs. 81.7%) (Fig. 3). We investigated the prognostic impact of TP53 mutations on MDS-EB and AML using survival analysis. In 10 patients with MDS-EB, shorter overall survival (OS) was associated with TP53 mutations (P=0.006; median OS 21.8 vs. 6.9 months) and multi-hit TP53 alterations (P=0.009; median OS, 21.8 vs. 6.9 months) (Supplemental Data Fig. S1), but not with the EB-1 or EB-2 subtype (P=0.159). Univariate analyses showed that multi-hit TP53 alterations, but not single TP53 mutation, had a significantly greater hazard ratio (HR) of death (HR=10.77, P=0.033). Survival analysis of the 37 AML patients revealed that TP53 mutations also had an adverse prognostic impact in AML patients, albeit with borderline significance (P=0.072; median OS 12.0 vs. 4.4 months) (Supplemental Data Fig. S1).
Using the 2022 WHO and 2022 ICC, we reclassified 15 (31.9%) and 27 (57.4%) out of 47 patients, respectively. This result was most likely a consequence of the addition of mutation-based definitions in the diagnostic criteria of AML, MR and the introduction of new entities, such as myeloid neoplasms with mutated TP53. The differences in the numbers of reclassified cases between the 2022 WHO and 2022 ICC categories were due to the inclusion of MR mutations in the previous category of MDS-EB2 and different entities of myeloid neoplasms with TP53 mutation. These new classifications improved our diagnostic capability of defining a highly adverse prognostic group, especially in patients with MDS with multi-hit TP53 alterations. With the increasing importance of detecting multi-hit TP53 alterations in the diagnostic approach in both the 2022 WHO and 2022 ICC, the adaptation of various molecular test methods may be necessary in clinical settings, especially in cases of multi-hit alterations with low VAFs [13]. In addition, although with borderline significance, we observed an adverse prognostic impact of TP53 mutations in patients with AML, suggesting that AML with mutated TP53 may be a distinct group with poor prognosis, consistent with the 2022 ICC [6].
In summary, the 2022 WHO and 2022 ICC identified different subtypes from previously heterogeneous diagnostic categories based on genetic characteristics. Changes in the blast thresholds for defining AML in some genetic abnormality categories were not associated with the major reclassified cases in the clinical setting.
Notes
AUTHOR CONTRIBUTIONS
Yoon SY and Yoon J contributed to the study conception and design; Kim HN, Kwon JA, Jeon MJ, Yu ES, Kim DS, and Choi CW were involved in clinical evaluation; Lee C, Yoon SY, and Yoon J interpreted the results; Lee C and Yoon J statistically analyzed the data; Lee C and Yoon J drafted the manuscript; and Yoon J supervised the study. All authors read and approved the final manuscript.
REFERENCES
1. Bernard E, Tuechler H, Greenberg PL, Hasserjian RP, Arango Ossa JE, Nannya Y, et al. 2022; Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evid. 1:EVIDoa2200008. DOI: 10.1056/EVIDoa2200008.

2. Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. 2022; Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 140:1345–77. DOI: 10.1182/blood.2022016867. PMID: 35797463.

3. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Cancer Genome Atlas Research Network. 2013; Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 368:2059–74. DOI: 10.1056/NEJMoa1301689. PMID: 23634996. PMCID: PMC3767041.

4. Park HS, Son SM, Kwon J. Serial analysis and comparison of mutation profiles in decitabine-treated myeloid sarcoma and subsequent acute myeloid leukemia using next-generation sequencing. Ann Lab Med. 2022; 42:602–5. DOI: 10.3343/alm.2022.42.5.602. PMID: 35470279. PMCID: PMC9057818.

5. Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. 2022; The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 36:1703–19. DOI: 10.1038/s41375-022-01613-1. PMID: 35732831. PMCID: PMC9252913.

6. Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. 2022; International Consensus Classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood. 140:1200–28. DOI: 10.1182/blood.2022015850. PMID: 35767897.
7. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. 2016; The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 127:2391–405. DOI: 10.1182/blood-2016-03-643544. PMID: 27069254.

8. McGowan-Jordan J, Hastings RJ, editors. 2020. ISCN 2020: An International System for Human Cytogenomic Nomenclature. Karger;Basel:
9. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. 2013; STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. DOI: 10.1093/bioinformatics/bts635. PMID: 23104886. PMCID: PMC3530905.

10. Arriba [Computer software]. (2018). ESMO Open, 3(Suppl 2), A179. https://github.com/suhrig/arriba. Updated on January 2022.
11. R [Computer software]. (2021). https://www.R-project.org/. Updated on November 2017.
12. Bolouri H, Farrar JE, Triche T Jr, Ries RE, Lim EL, Alonzo TA, et al. 2018; The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 24:103–12. DOI: 10.1038/nm.4439. PMID: 29227476. PMCID: PMC5907936.

13. Bernard E, Nannya Y, Hasserjian RP, Devlin SM, Tuechler H, Medina-Martinez JS, et al. 2020; Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med. 26:1549–56. DOI: 10.1038/s41591-020-1008-z. PMID: 32747829. PMCID: PMC8381722.

14. Grob T, Al Hinai ASA, Sanders MA, Kavelaars FG, Rijken M, Gradowska PL, et al. 2022; Molecular characterization of mutant TP53 acute myeloid leukemia and high-risk myelodysplastic syndrome. Blood. 139:2347–54. DOI: 10.1182/blood.2021014472. PMID: 35108372.
15. Haase D, Stevenson KE, Neuberg D, Maciejewski JP, Nazha A, Sekeres MA, et al. 2019; TP53 mutation status divides myelodysplastic syndromes with complex karyotypes into distinct prognostic subgroups. Leukemia. 33:1747–58. DOI: 10.1038/s41375-018-0351-2. PMID: 30635634. PMCID: PMC6609480.

Fig. 1
Summary of the reclassification of the study population according to the 2022 WHO and 2022 ICC classifications. *A case diagnosed as therapy-related myeloid neoplasm according to the 2016 WHO classification is not included in the above figure. It was reclassified as AML, MR post cytotoxic therapy and AML with mutated TP53, therapy-related according to the 2022 WHO and 2022 ICC classifications, respectively.
Abbreviations: MDS-EB, MDS with excess blasts; ICC, International Consensus Classification; IB, increased blasts; MR, myelodysplasia-related; MRC, myelodysplasia-related changes; NOS, not otherwise specified.
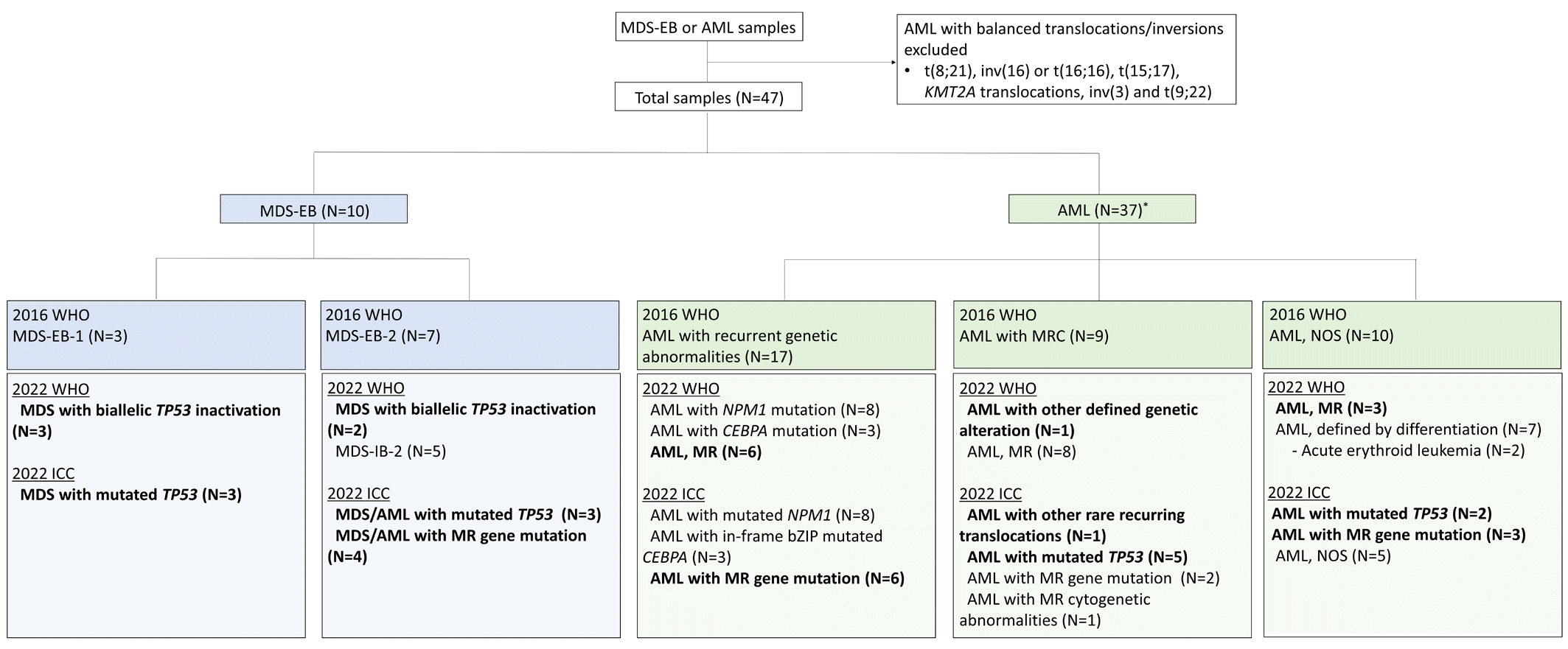
Fig. 2
Relationships between subtypes in the study population classified according to the 2016 WHO, 2022 WHO, and 2022 ICC classifications. Reclassification based on the (A) 2022 WHO and (B) 2022 ICC classifications. *AML with recurrent genetic abnormalities is not a valid category in the 2022 ICC classification. The term was adopted from the WHO to characterize eight AML cases with mutated NPM1, three AML cases with in-frame bZIP-mutated CEBPA, and one AML case with other rare recurring translocations.
Abbreviations: MDS-EB, MDS with excess blasts; MRC, myelodysplasia-related changes; NOS, not otherwise specified; t-MN, therapy-related myeloid neoplasms; IB, increased blasts; MR, myelodysplasia-related; ICC, International Consensus Classification.
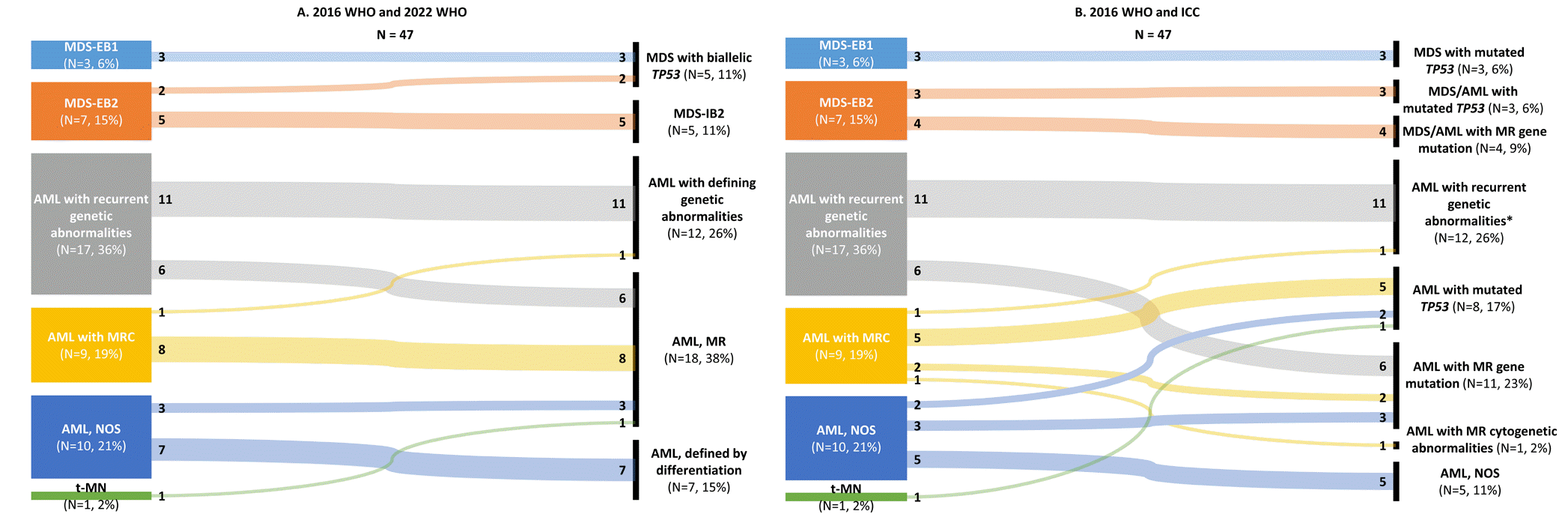
Fig. 3
Boxplots showing TP53 VAF levels stratified by TP53 mutation states. (A) Comparison of VAF levels between the single TP53 mutation (1mut) and multi-hit TP53 alteration groups. (B) Comparison of VAF levels between single TP53 mutation and various subtypes of multi-hit TP53 alterations. Multi-hit TP53 alterations comprise two TP53 mutations (2mut), a single TP53 mutation with copy number loss of TP53 (Mut+del), and a single TP53 mutation with cnLOH (Mut+cnLOH). *P<0.1, Wilcoxon–Mann–Whitney test; each multi-hit TP53 alteration subgroup was compared to the single TP53 mutation group.
Abbreviations: VAF, variant allele fraction; cnLOH, copy number neutral loss of heterozygosity.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download