Introduction
Materials and Methods
1. Patient and study procedure
2. Tumor sample collection for whole-exome and transcriptome sequencing
3. Statistical analysis
Results
1. Clinical and pathological characteristics of sarcoma patients
Table 1
| Variable | Total | Pazopanib | Pazopanib/Durvalumab | p-valuec) |
|---|---|---|---|---|
| No. of patients | 35 | 18 (51.4) | 17 (48.6) | |
| Age (yr) | 48 | 51 (38–73) | 45 (22–72) | |
| Sex | ||||
| Male | 16 | 8 (50.0) | 8 (50.0) | 0.78 |
| Female | 19 | 10 (52.6) | 9 (47.4) | |
| ECOG PS | ||||
| 0 | 10 | 3 (30.0) | 7 (70.0) | 0.14* |
| 1 | 25 | 15 (60.0) | 10 (40.0) | |
| Histologic variant | ||||
| LMS | 8 | 6 (75.0) | 2 (25.0) | 0.23* |
| UPS | 7 | 5 (71.4) | 2 (28.6) | |
| MPNST | 5 | 1 (20.0) | 4 (80.0) | |
| DDLPS | 5 | 3 (60.0) | 2 (40.0) | |
| Synovial sarcoma | 4 | 1 (25.0) | 3 (75.0) | |
| Othersa) | 6 | 2 (33.3) | 4 (66.7) | |
| Primary site | ||||
| Upper abdomen/Retroperitoneum | 11 | 8 (72.7) | 3 (27.3) | 0.10* |
| Extremity | 6 | 0 | 6 (100) | |
| Gynecologic organ | 6 | 3 (50.0) | 3 (50.0) | |
| Lower abdomen | 5 | 4 (80.0) | 1 (20.0) | |
| Head/Neck | 5 | 2 (40.0) | 3 (60.0) | |
| Thorax | 2 | 1 (50.0) | 1 (50.0) | |
| Stage at diagnosisb) | ||||
| II | 15 | 6 (40.0) | 9 (60.0) | 0.42* |
| III | 14 | 9 (64.3) | 5 (35.7) | |
| IV | 6 | 3 (50.0) | 3 (50.0) | |
| FNCLCC grade | ||||
| I | 4 | 4 (100) | 0 | 0.10* |
| II | 8 | 3 (37.5) | 5 (62.5) | |
| III | 23 | 11 (47.8) | 12 (52.2) | |
| No. of previous chemotherapy regimens | ||||
| 1 | 17 | 6 (35.3) | 11 (64.7) | 0.21* |
| 2 | 12 | 7 (58.3) | 5 (41.7) | |
| > 3 | 6 | 5 (83.3) | 1 (16.7) | |
| Sequencing samples | ||||
| Primary tumor | 16 | 6 (37.5) | 10 (62.5) | 0.13 |
| Metastatic sites | 19 | 12 (63.2) | 7 (36.8) | |
| Type of previous chemotherapy received | ||||
| Doxorubicin monotherapy | 9 | 8 (72.8) | 1 (11.1) | NA |
| Gemcitabine/Docetaxel | 11 | 6 (54.5) | 5 (45.5) | |
| Eribulin | 1 | 1 (100) | 0 | |
| Doxorubicin/Ifosfamide | 9 | 3 (33.3) | 6 (66.6) | |
| Doxorubicin/Olaratumab | 13 | 6 (46.2) | 7 (53.8) | |
| Ifosfamide combination | 15 | 11 (73.3) | 4 (26.7) | |
Values are presented as number (%) or median (range). DDLPS, dedifferentiated liposarcoma; ECOG PS, Eastern Cooperative Oncology Group performance status; FNCLCC, Fédération Nationale des Centres de Lutte Contre le Cancer; LMS, leiomyosarcoma; MPNST, malignant peripheral nerve sheath tumor; NA, not available; UPS, undifferentiated pleomomorphic sarcoma.
a) Others: desmoplastic small round cell tumor, myxoid fibrosarcoma, high grade endometrial stromal cell sarcoma, clear cell sarcoma, low grade myofibroblastic sarcoma, and solitary fibrous tumor,
2. Molecular characterization and response to pazopanib-based treatment
Fig. 1
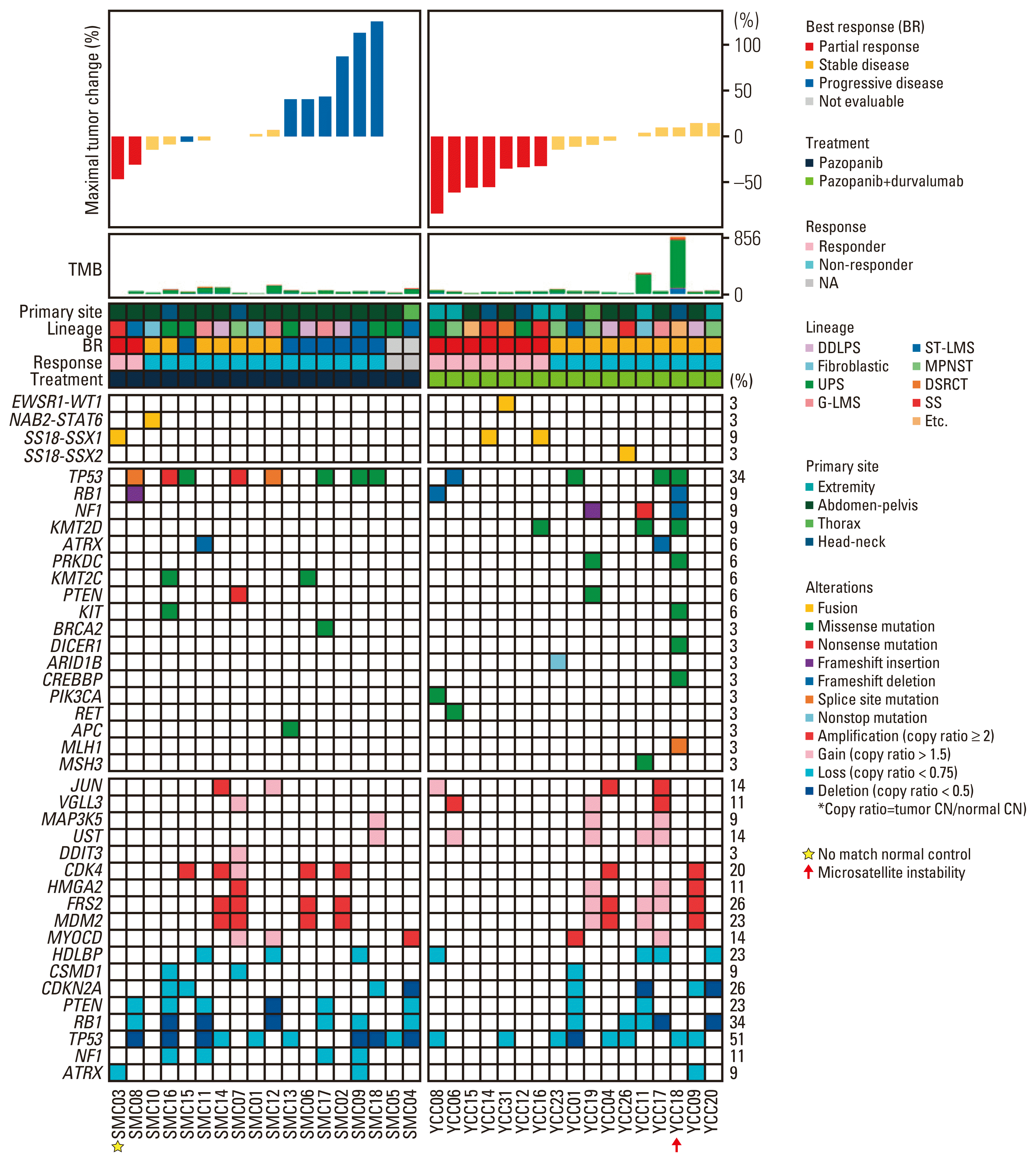
Fig. 2
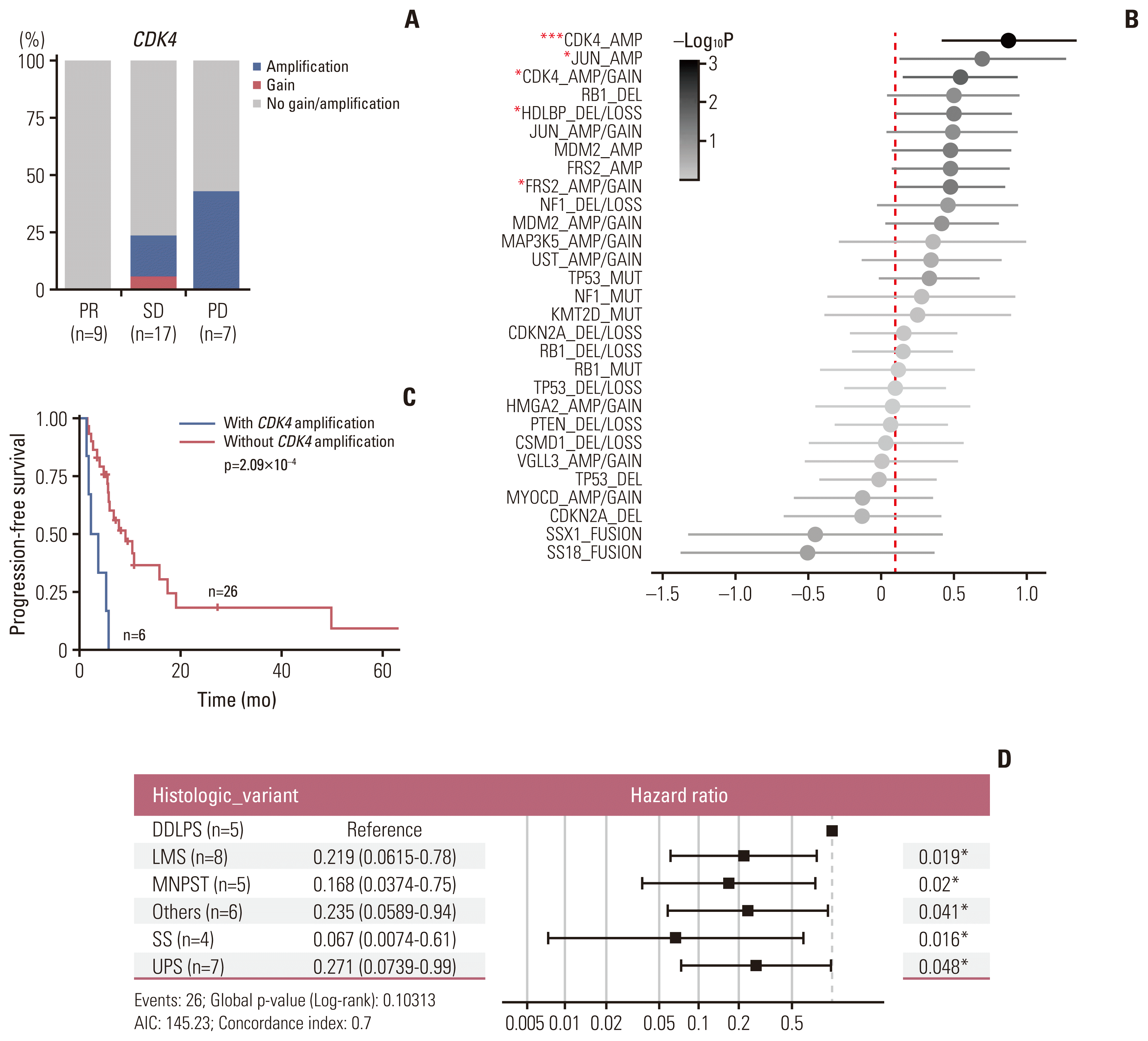
3. Genomic landscape and correlates of pazopanib efficacy
Fig. 3
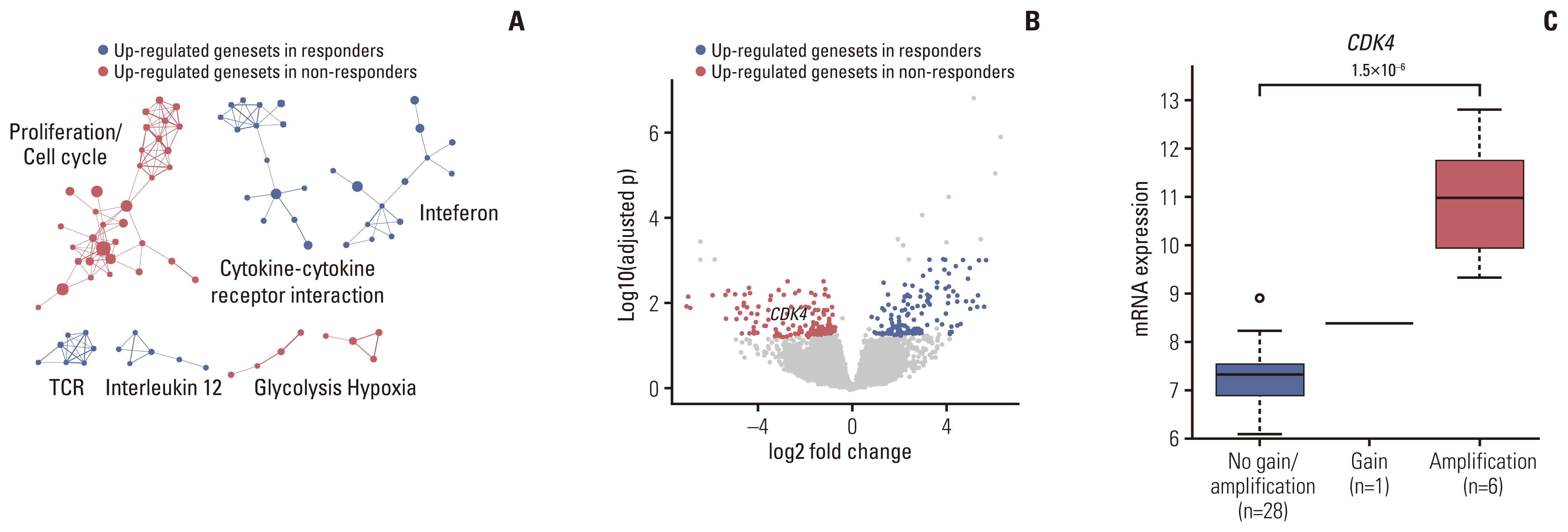
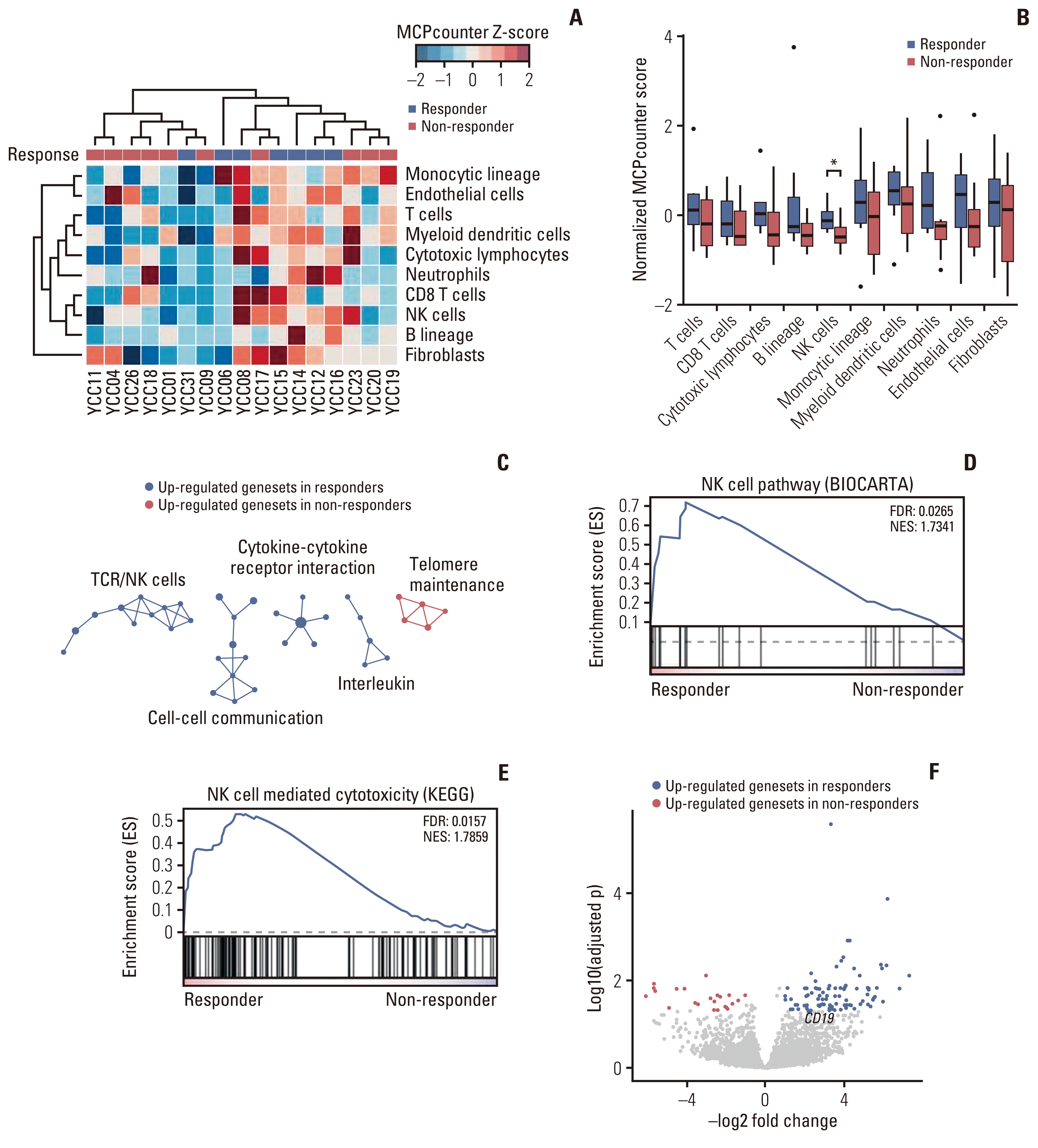
4. Identification of transcriptional determinants dictating clinical response to pazopanib-based treatment
Fig. 4
5. Predictors of treatment response to pazopanib and programmed death-ligand 1 blockade combination
Fig. 5




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download