1. Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996; 87:159–170. PMID:
8861899.
2. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002; 16:6–21. PMID:
11782440.

3. Russo VEA, Martienssen RA, Riggs AD. Epigenetic mechanisms of gene regulation. New York: Cold Spring Harbor Laboratory Press;1996.
4. Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999; 21:163–167. PMID:
9988266.
5. Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003; 349:2042–2054. PMID:
14627790.

6. Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007; 8:286–298. PMID:
17339880.
7. Esteller M. Epigenetics in cancer. N Engl J Med. 2008; 358:1148–1159. PMID:
18337604.

8. Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferaselike protein DNMT3L stimulates
de novo methylation by Dnmt3a. Proc Natl Acad Sci U S A. 2002; 99:16916–16921. PMID:
12481029.
9. Deplus R, Brenner C, Burgers WA, et al. Dnmt3L is a transcriptional repressor that recruits histone deacetylase. Nucleic Acids Res. 2002; 30:3831–3838. PMID:
12202768.

10. Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007; 128:683–692. PMID:
17320506.

11. Wajed SA, Laird PW, DeMeester TR. DNA methylation: an alternative pathway to cancer. Ann Surg. 2001; 234:10–20. PMID:
11420478.
12. Gu M, Kim D, Bae Y, Choi J, Kim S, Song S. Analysis of microsatellite instability, protein expression and methylation status of hMLH1 and hMSH2 genes in gastric carcinomas. Hepatogastroenterology. 2009; 56:899–904. PMID:
19621725.
13. Kane MF, Loda M, Gaida GM, et al. Methylation of the
hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997; 57:808–811. PMID:
9041175.
14. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996; 93:9821–9826. PMID:
8790415.

15. Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000; 92:564–569. PMID:
10749912.
16. Merlo A, Herman JG, Mao L, et al. 5'CpG island methylation is associated with transcriptional silencing of the tumour suppressor
p16/CDKN2/MTS1 in human cancers. Nat Med. 1995; 1:686–692. PMID:
7585152.
17. Greger V, Passarge E, Hopping W, Messmer E, Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet. 1989; 83:155–158. PMID:
2550354.

18. Chan TA, Glockner S, Yi JM, et al. Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Med. doi:
10.1371/journal.pmed.0050114. Published online 27 May 2008.

19. Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007; 3:1709–1723. PMID:
17892325.

20. Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983; 301:89–92. PMID:
6185846.

21. Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002; 99:3740–3745. PMID:
11891299.
22. Bird A. The essentials of DNA methylation. Cell. 1992; 70:5–8. PMID:
1377983.

23. Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003; 300:455. PMID:
12702868.

24. Rodriguez J, Frigola J, Vendrell E, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006; 66:8462–9468. PMID:
16951157.

25. Fraga MF, Herranz M, Espada J, et al. A mouse skin multistage carcinogenesis model reflects the aberrant DNA methylation patterns of human tumors. Cancer Res. 2004; 64:5527–5534. PMID:
15313885.

26. Antequera F, Bird A. CpG islands. EXS. 1993; 64:169–185. PMID:
8418949.
27. Lim DH, Maher ER. Genomic imprinting syndromes and cancer. Adv Genet. 2010; 70:145–175. PMID:
20920748.

28. Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010; 70:27–56. PMID:
20920744.

29. Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010; 28:573–621. PMID:
20192811.

30. Loftus EV Jr. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004; 126:1504–1517. PMID:
15168363.

31. Thia KT, Loftus EV Jr. SandbornWJ. YangSK. An update on the epidemiology of inflammatory bowel disease in Asia. Am J Gastroenterol. 2008; 103:3167–3182. PMID:
19086963.
32. Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001; 293:1068–1070. PMID:
11498573.

33. Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007; 448:427–434. PMID:
17653185.

34. Kim JM. Antimicrobial proteins in intestine and inflammatory bowel diseases. Intest Res. 2014; 12:20–33. PMID:
25349560.

35. Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012; 491:119–124. PMID:
23128233.
36. Xavier RJ, Rioux JD. Genome-wide association studies: a new window into immune-mediated diseases. Nat Rev Immunol. 2008; 8:631–643. PMID:
18654571.
37. Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008; 40:955–962. PMID:
18587394.
38. Bird AP, Wolffe AP. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999; 99:451–454. PMID:
10589672.

39. Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010; 465:721–727. PMID:
20535201.

40. Gloria L, Cravo M, Pinto A, et al. DNA hypomethylation and proliferative activity are increased in the rectal mucosa of patients with long-standing ulcerative colitis. Cancer. 1996; 78:2300–2306. PMID:
8940998.

41. Hsieh CJ, Klump B, Holzmann K, Borchard F, Gregor M, Porschen R. Hypermethylation of the p16
INK4a promoter in colectomy specimens of patients with long-standing and extensive ulcerative colitis. Cancer Res. 1998; 58:3942–3945. PMID:
9731506.
42. Tahara T, Shibata T, Nakamura M, et al. Effect of MDR1 gene promoter methylation in patients with ulcerative colitis. Int J Mol Med. 2009; 23:521–527. PMID:
19288029.

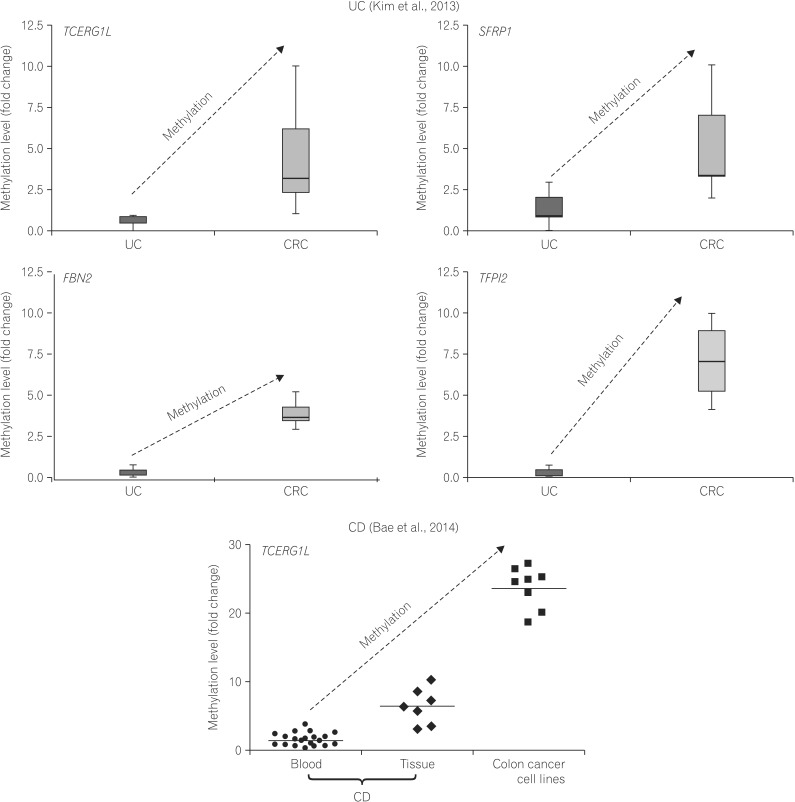
43. Kim TO, Park J, Kang MJ, et al. DNA hypermethylation of a selective gene panel as a risk marker for colon cancer in patients with ulcerative colitis. Int J Mol Med. 2013; 31:1255–1261. PMID:
23546389.

44. Lin Z, Hegarty JP, Cappel JA, et al. Identification of diseaseassociated DNA methylation in intestinal tissues from patients with inflammatory bowel disease. Clin Genet. 2011; 80:59–67. PMID:
20950376.

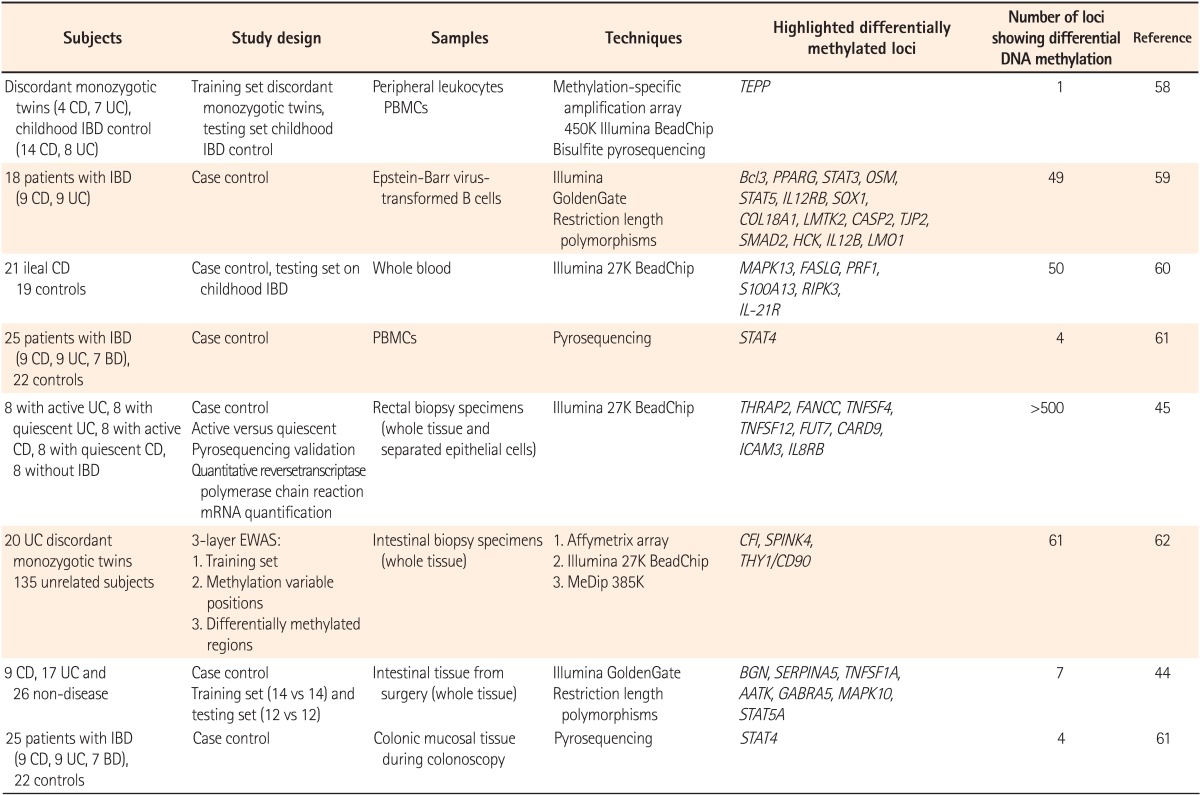
45. Cooke J, Zhang H, Greger L, et al. Mucosal genome-wide methylation changes in inflammatory bowel disease. Inflamm Bowel Dis. 2012; 18:2128–2137. PMID:
22419656.

46. Saito S, Kato J, Hiraoka S, et al. DNA methylation of colon mucosa in ulcerative colitis patients: correlation with inflammatory status. Inflamm Bowel Dis. 2011; 17:1955–1965. PMID:
21830274.
47. Bae JH, Park J, Yang KM, Kim TO, Yi JM. Detection of DNA hypermethylation in sera of patients with Crohn's disease. Mol Med Rep. 2014; 9:725–729. PMID:
24317008.

48. Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000; 28:e32. PMID:
10734209.

49. Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002; 23:4072–4079. PMID:
12481262.

50. Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008; 452:553–563. PMID:
18385729.

51. Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009; 100:2014–2021. PMID:
19719775.
52. Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003; 3:253–266. PMID:
12671664.

53. Glockner SC, Dhir M, Yi JM, et al. Methylation of
TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009; 69:4691–4699. PMID:
19435926.
54. Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008; 54:414–423. PMID:
18089654.

55. Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008; 358:1118–1128. PMID:
18337602.

56. Yi JM, Dhir M, Van Neste L, et al. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin Cancer Res. 2011; 17:1535–1545. PMID:
21278247.
57. Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999; 59:793–797. PMID:
10029064.
58. Harris RA, Nagy-Szakal D, Pedersen N, et al. Genome-wide peripheral blood leukocyte DNA methylation microarrays identified a single association with inflammatory bowel diseases. Inflamm Bowel Dis. 2012; 18:2334–2341. PMID:
22467598.

59. Lin Z, Hegarty JP, Yu W, et al. Identification of disease-associated DNA methylation in B cells from Crohn's disease and ulcerative colitis patients. Dig Dis Sci. 2012; 57:3145–3153. PMID:
22821069.

60. Nimmo ER, Prendergast JG, Aldhous MC, et al. Genome-wide methylation profiling in Crohn's disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm Bowel Dis. 2012; 18:889–899. PMID:
22021194.

61. Kim SW, Kim ES, Moon CM, Kim TI, Kim WH, Cheon JH. Abnormal genetic and epigenetic changes in signal transducer and activator of transcription 4 in the pathogenesis of inflammatory bowel diseases. Dig Dis Sci. 2012; 57:2600–2607. PMID:
22569826.
62. Hasler R, Feng Z, Backdahl L, et al. A functional methylome map of ulcerative colitis. Genome Res. 2012; 22:2130–2137. PMID:
22826509.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download