Abstract
The ABO system remains the most important blood group classification system in transfusion medicine. ABO serology testing requires both forward and reverse typing as each grouping result validates the other, and ABO discrepancies must be resolved in the clinical laboratory. ABO genotyping has served as an independent tool in accurate blood group determination. This report retrospectively analyzes the ABO grouping results of 47 blood samples from two medical institutions. Various genotype-phenotype results were categorized based on correlations reported by previous investigators, and patients’ medical records were reviewed for conditions that may have affected the results. The frequencies of genotype-phenotype matches and genotype-phenotype mismatches were 72.3% (34/47 cases) and 27.7% (13/47cases), respectively. The cis-AB alleles (23 cases) were the most prevalent cause of ABO discrepancies. Red cell A or B antigen losses in patients with hematologic disorders, malignancy, and pregnancy were genetically confirmed. A pedigree study revealed a father-son pair with the same genotype showing differing phenotypes, and whole exon sequencing including promoter region revealed a single-point mutation in the promoter region in one of the two patients. By directing sequencing of the full ABO coding region, including the promoter, in the analysis, we aimed to determine the potential regulatory role of a mutation in the promoter region in ABO gene expression.
초록
안전한 수혈을 위해서는 ABO 혈액형 검사가 중요하며, 혈청학적 검사 소견의 불일치 해결 및 ABO 아형 감별에 있어, ABO 유전형 검사가 유용하게 사용된다. 본 연구는 두 개의 의료기관에서 47명의 환자들에게 시행한 ABO 혈청학적 검사와 유전형 검사 결과를 후향적으로 분석하였다. ABO 유전형은 PCR 증폭 및 직접염기서열분석법을 통해 분석하였고, 환자의 질병 상태 파악을 위한 의무 기록 또한 검토하였다. 분석 결과, 유전형-표현형 일치와 유전형-표현형 불일치 예가 각각 72.3% (34/47예)와 27.7% (13/47예)의 빈도를 보였다. Cis-AB 대립유전자(23예)에 의한 ABO 불일치가 가장 높은 비율을 차지하였으며 혈액질환, 악성종양, 임신과 관련된 환자들에게서 적혈구 A 또는 B 항원 소실이 유전적으로 확인되었다. 또한 가계도 조사에서 동일한 cis-AB.01/A1.02 유전형을 보였으나 다른 표현형을 보인 부자 한 쌍에 대하여 프로모터를 포함한 모든 엑손 부위에 대해 추가적으로 염기서열분석을 진행하였고, 그 결과, 프로모터 영역의 단일염기다형성에 차이가 있음을 발견하였다. 유전자 발현에 있어 프로모터 영역의 잠재적인 조절 역할에 대한 추가적인 평가가 필요할 것으로 생각된다.
The ABO blood group shows phenotypic variants, largely owing to its molecular genetic basis [1]. As an independent tool in the clinical laboratory, serology can reveal ABO discrepancies and ABO genotyping is essential for correct determination of the blood group. The cis-AB blood group largely explains the ABO discrepancies in the East-Asian population [2]. The cis-AB allele results in a mutated enzyme with both A and B glycosyltransferase activities and leads to decreased levels of both antigens, which is presented as various phenotypes [3-5]. The first phenotypes identified were A2B3, A2B, and A1B3, which are associated with the cis-AB01/O, cis-AB01/B, and cis-AB01/A genotypes, respectively [6]. Further studies revealed more phenotypes, including A1Bel, A1Bx, A1Bm, AintB3, AintB, and A1 [7].
In this study, we performed a retrospective investigation of the ABO grouping test results of two medical institutions in Korea. When weak or unusual expression of antigens was observed, the ABO genotyping results and medical records were evaluated. In selected samples for which the cause of ABO discrepancy remained unresolved, we looked for genetic variants in the full ABO coding region and promoter region. We reviewed the serology and genotyping results of 39 samples from Seoul St. Mary’s Hospital and 8 samples from Incheon St. Mary’s Hospital from January 2016 to June 2021. Additionally, we included pedigree analyses of two families. This study was approved by the Institutional Review Board (IRB) of the Catholic Medical Center (Seoul, Korea) and the need for informed consent was waived (XC22RADI0073).
Serological tests were routinely performed using a column agglutination technique using ORTHO BioVue System (Ortho Clinical Diagnostics, Pencoed, UK) on an automatic analyzer, ORTHO VISION (Ortho Clinical Diagnostics, Raritan, NJ, USA). In the presence of weak or unusual expression of the A or B antigens, manual tube methods using murine monoclonal anti-A, anti-B (Shinyang Diagnostics, Seoul, Korea) were employed, and serum typing was done with A1 and B cells (MIRR. SciTech Corp., Seoul, Korea) using a plate method. To further identify ABO subgroups, anti-A1 lectin obtained from Dolichos biflorus seeds and anti-H from Ulex europaeus seeds (Lorne Laboratories Ltd, Berkshire, UK) were used.
For ABO genotyping, DNA was extracted from EDTA blood samples using a QIAsymphony DSP DNA Kit (Qiagen, Hilden, Germany). PCR was run using a C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA, USA) with a mixture of primer pairs (Table 1) for amplification. The products were purified using the ExoSAP-IT PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, MA, USA). A Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and an ABI 3500XL DX Genetic Analyzer (Applied Biosystems) were used for Sanger sequencing according to the manufacturer’s instructions. Sequencing data were analyzed using the ABO reference sequence (NM_020469.2) with Sequencher v.5 (Gene Codes Corp, Ann Arbor, MI, USA). The ABO alleles were classified according to the nomenclature used by the International Society of Blood Transfusion.
A total of 47 ABO serology and genotyping results were reviewed. These were referred for ABO genotyping test as they all showed a ABO discrepancy—defined as a mismatch between the cell and serum typing or presence of a weaker strength of agglutination (≤3+ in cell typing; ≤2+ in serum typing), as described in a previous study [8]. Results were further classified as ‘phenotype-genotype matches’ or ‘phenotype-genotype mismatches’ (Table 2). The frequency of phenotypes consistent with their genotypes was 72.3% (34/47 cases). Among them, 23 cases demonstrated a cis-AB allele by genotyping, with cis-AB/O being the most common genotype. All samples with A2B3 phenotype had cis-AB/O alleles but not necessarily vice versa. Cis-AB.01/O.01.01 and cis-AB.01/O.01.02 genotypes were also associated with A1B3 or AB phenotypes. All cis-AB.01/B.01 patients expressed A1B3 phenotypes. Consistent with previous studies, two patients with cis-AB.01/A1.02 genotype demonstrated A phenotype. One patient with cis-AB.01/A1.02 genotype did not show agglutination with anti-B in cell typing but showed weak agglutination with B cell in serum typing, and was thus categorized as A1Bel. Other weak ABO subgroups included Ax/Aweak, an uncommon phenotype with a 0.01% frequency in a large-scale study [9].
The frequency of phenotypes inconsistent with their genotypes was 27.7% (13/47 cases). Serology showed five distinct A subgroups (A2, A3, Aw, Ael, Aint), but genotypes of all samples included the common A1.02 allele. Direct sequencing of exons 3 to 5 was performed for a patient who showed Ael phenotype but A1.02/O.01.01 genotype, and no other variants were detected. Whole exon sequencing including the promoter region was recommended to the clinical department. The serology result of another patient showed AwB, but her genotype was A1.02/B.01. This discrepancy was confirmed by pedigree analysis of her father’s and mother’s genotyping results (Fig. 1B). The patient continued to show weakened A antigen expression in follow-up testing. Furthermore, while two different B subgroups (B3, Bw) were expressed, 6 out of 7 patients had the consensus B.01 allele. One patient with Bw phenotype initially showed 6 possible genotypes in sequencing. Further analysis of exons 3 to 5 showed additional variants, and the patient’s genotype was narrowed down to either B.01/O.01.02 or B3.02/O.01.35. Further studies such as allele specific sequencing and family studies could have located the allele that contained the variants.
Medical records were evaluated for any medical factors that may have affected the test results. Weak but variable A or B antigen expression was noted in patients with hematological diseases (N=5), including juvenile myelomonocytic leukemia, Burkitt lymphoma, high-grade B-cell lymphoma, acute myeloid leukemia, and myelodysplastic syndrome. All patients showed weak antigen expression at initial diagnosis, and none of them received out-of-group transfusion prior to serological typing. Interestingly, one patient with myelodysplastic syndrome evolved into leukemic transformation 9 months after initial diagnosis. One patient who showed weak B antigen expression was diagnosed with renal cell carcinoma.
Pregnancy was suspected to cause weak antigen expression in 3 patients. Notably, one multipara, who showed normal AB blood grouping results, exhibited weak A antigen expression (3+ in cell typing, +/- in serum typing) during her third pregnancy. When ABO genotyping was performed, she was diagnosed as cis-AB.01/B.01. Repeating sample testing 4 years after her third delivery expressed a normal AB. This patient showed a simultaneous presence of medical factor and weak ABO subgroup allele. Other women were unattainable for sample testing to be repeated after delivery.
Pedigree analysis was performed for one family that showed cis-AB inheritance across three generations. Interestingly, both the father and paternal grandfather had cis-AB.01/A1.02 genotype with different phenotypes: A1Bel, A (Fig. 1A), and the results were the same after retesting. Therefore, whole exon sequencing including promoter region was performed, and a single-point mutation in the promoter region (-219 C>A) was found in one of the two patients (Fig. 2). In transferase analysis, the patient who had a mutation in the promoter region had A transferase but not B transferase, which led to A phenotype. The other patient who did not possess the mutation had both A and B transferases and presented as A1Bel. We suggest that the expression of the ABO gene was suppressed by the mutation of the promoter region, resulting in A phenotype even though the patient’s genotype was cis-AB.01/A1.02. This in turn implies that the single point mutation was on the cis-AB-allele in the individual.
The ABO gene, located on chromosome 9q34, consists of seven coding exons, with the largest open reading frame located in exons 6 and 7 [10]. As analysis of only exons 6 and 7 is sometimes insufficient, further sequencing of other exons, introns, and the promoter region is required [11, 12]. In 1997, Kominato et al. first verified the essential role of the promoter sequence in the regulation of ABO gene transcription using KATO III, the human gastric cancer cell line [13, 14]. Recent studies revealed cases where mutations of the promoter sequence downregulate the promoter activity, thereby decreasing A or B antigen expression. Isa et al. [15] discovered two single-point mutations (-76G>C and -86G>T) in the promoter on the A-allele in three A3 patients with A1.01/O.01.01 and A1.01/O.01.02 as well as on the B-allele in a B3 patient with B.01/O.01.02. These findings were verified using a luciferase assay that compared the promoter activity of reporter plasmids carrying the promoters with mutations in the -76 and -68 regions with that of a wild type promoter [15]. Additionally, Hellberg et al. [16] reported a mutation in the -72 region in a patient who showed phenotype B3 but B.01/O.01.01 genotype.
In conclusion, the prevalence of the ABO subgroups demonstrated in this study was consistent with that of similar previously published studies. Medical factors, including hematologic disorders, malignancy, and pregnancy also affected the ABO discrepancies, as noted previously [17]. This study further aimed to focus on the effect of a mutation in the promoter region on ABO gene expression. Further evaluation of the promoter region could help differentiate ABO discrepancies that remain unresolved.
REFERENCES
1. Ogasawara K, Yabe R, Uchikawa M, Saitou N, Bannai M, Nakata K, et al. 1996; Molecular genetic analysis of variant phenotypes of the ABO blood group system. Blood. 88:2732–7. DOI: 10.1182/blood.V88.7.2732.bloodjournal8872732. PMID: 8839869.
2. Cho D, Kim SH, Jeon MJ, Choi KL, Kee SJ, Shin MG, et al. 2004; The serological and genetic basis of the cis-AB blood group in Korea. Vox Sang. 87:41–3. DOI: 10.1111/j.1423-0410.2004.00528.x. PMID: 15260821.
3. Yoshida A. 1981; Genetic mechanism of blood group (ABO)-expression. Acta Biol Med Ger. 40:927–41.
4. Yoshida A, Yamaguchi H, Okubo Y. 1980; Genetic mechanism of cis-AB inheritance. II. Cases associated with structural mutation of blood group glycosyltransferase. Am J Hum Genet. 32:645–50.
5. Yoshida A, Yamaguchi H, Okubo Y. 1980; Genetic mechanism of cis-AB inheritance. I. A case associated with unequal chromosomal crossing over. Am J Hum Genet. 32:332–8.
6. Yamaguchi H. 1973; A review of cis AB blood. Jinrui Idengaku Zasshi. 18:1–9.
7. Chun S, Choi S, Yu H, Cho D. 2019; Cis-AB, the blood group of many faces, is a conundrum to the novice eye. Ann Lab Med. 39:115–20. DOI: 10.3343/alm.2019.39.2.115. PMID: 30430772. PMCID: PMC6240514.
8. Heo WY, Chung YN, Kim TY, Yu H, Bae JC, Kim H, et al. 2021; Analysis of ABO grouping discrepancies among patients from a tertiary hospital in Korea. Transfus Apher Sci. 60:103230. DOI: 10.1016/j.transci.2021.103230. PMID: 34400096.
9. Anukul N, Leetrakool N, Tanan P, Palacajornsuk P, Klangsinsirikul P. 2018; Mixed-field agglutination observed in column agglutination testing is not always associated with the A3 subgroup. Immunohematology. 34:49–56. DOI: 10.21307/immunohematology-2018-009. PMID: 29989419.
10. Olsson ML, Westman JS. Cohn CS, Delaney M, editors. 2020. The ABO system (001). Technical manual. 20th ed. p. 302. Bethesda, MD: Association for the Advancement of Blood & Biotherapies.
11. Yu LC, Twu YC, Chou ML, Chang CY, Wu CY, Lin M. 2002; Molecular genetic analysis for the B3 allele. Blood. 100:1490–2. DOI: 10.1182/blood-2002-01-0188. PMID: 12149236.
12. Seltsam A, Das Gupta C, Bade-Doeding C, Blasczyk R. 2006; A weak blood group A phenotype caused by a translation-initiator mutation in the ABO gene. Transfusion. 46:434–40. DOI: 10.1111/j.1537-2995.2006.00740.x. PMID: 16533287.
13. Kominato Y, Tsuchiya T, Hata N, Takizawa H, Yamamoto F. 1997; Transcription of human ABO histo-blood group genes is dependent upon binding of transcription factor CBF/NF-Y to minisatellite sequence. J Biol Chem. 272:25890–8. DOI: 10.1074/jbc.272.41.25890. PMID: 9325321.
14. Olsson ML, Chester MA. 2001; Polymorphism and recombination events at the ABO locus: a major challenge for genomic ABO blood grouping strategies. Transfus Med. 11:295–313. DOI: 10.1046/j.1365-3148.2001.00320.x. PMID: 11532186.
15. Isa K, Yamamuro Y, Ogasawara K, Yabe R, Ogiyama Y, Ito S, et al. 2016; Presence of nucleotide substitutions in the ABO promoter in individuals with phenotypes A3 and B3. Vox Sang. 110:285–7. DOI: 10.1111/vox.12363. PMID: 26529276.
16. Hellberg A, Hult AK, Moser I, Tomaz B, Rodrigues M, Olsson ML. 2019; A novel single-nucleotide substitution in the proximal ABO promoter gives rise to the B3 phenotype. Transfusion. 59:E1–3. DOI: 10.1111/trf.15457.
17. Olsson ML, Irshaid NM, Hosseini-Maaf B, Hellberg A, Moulds MK, Sareneva H, et al. 2001; Genomic analysis of clinical samples with serologic ABO blood grouping discrepancies: identification of 15 novel A and B subgroup alleles. Blood. 98:1585–93. DOI: 10.1182/blood.V98.5.1585. PMID: 11520811.
Fig. 1
Pedigree of the family 1 and 2. Arrow indicates propositus. (A) Pedigree of family 1: ABO phenotypes, genotypes, and transferase results are noted. (B) Pedigree of family 2: ABO phenotypes and genotypes are noted.
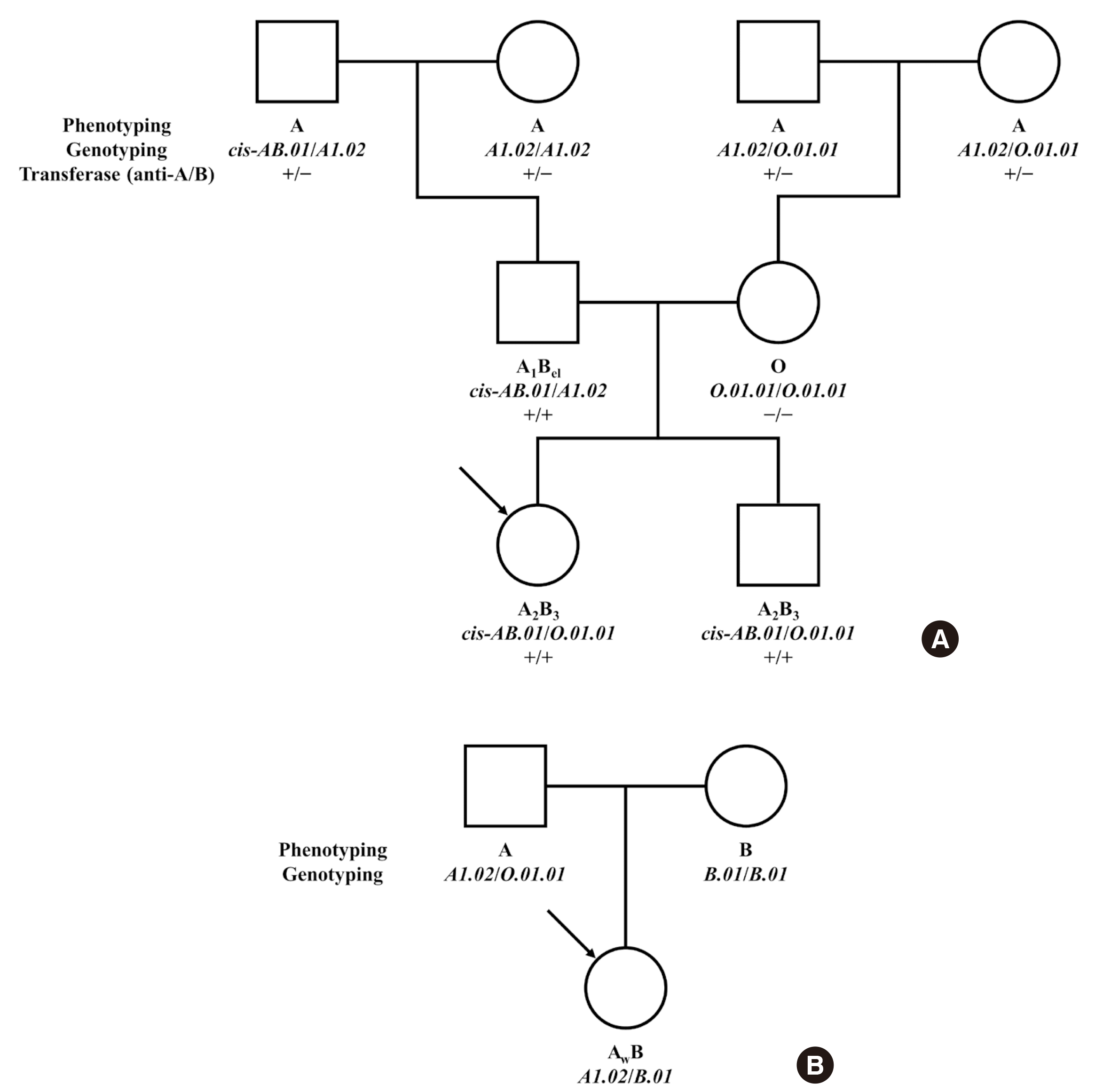
Fig. 2
Sequencing results of the promoter region of the ABO gene. The arrow indicates the mutation site. (A) Grandfather (p) shows a mutation at -219C>A. (B) Father does not have a mutation.

Table 1
Primers used in the study for Sanger sequencing of the promoter and whole exon of the ABO gene
Table 2
Phenotypic and genotypic results of patients in this study




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download