Abstract
Background
Coffin-Siris syndrome (CSS) is a rare disease characterized by features such as developmental delay, intellectual disability, unique facial feature, hypoplasia of the fifth finger or toe, hypertrichosis, and sparse scalp hair. CSS is currently diagnosed through a molecular genetic test that detects heterozygous pathogenic variants and deletion/duplication in the causative genes.
Methods
We retrospectively reviewed the medical records of 23 suspected patients with CSS enrolled in the rare disease diagnostic program of the Korean Disease Control and Prevention Agency from January 2017 to December 2020, including whole-exome sequencing (WES) reports. Statistical analysis was performed using cluster analysis through Jaccard/Tanimoto similarity test using the R version 4.2.0.
Results
Eight cases were genetically diagnosed with the CSS. Five cases were identified to have a novel variant: ARID1B (NM_020732.3) Gln958*, Asn1320*, Gly1696*, Gly806Trpfs*, and SMARCA4 (NM_001128849.1) Asn916Ser. Central nervous system symptoms were observed in all ARID1B cases, and the fifth digit hypoplasia was observed in all SMARCA4 cases. SMARCA4 Asn916Ser was identified as de novo. A similarity network was identified using cluster analysis, a relatively fresh approach to genotype-phenotype analysis.
Conclusions
We reported eight patients diagnosed with CSS, five of whom have novel genetic variants of ARID1B or SMARCA4. A novel case of SMARCA4 was de novo. This study contributes to describing the CSS phenotype. Future studies may facilitate easier diagnosis of CSS in patients who present with atypical traits as more in-depth genetic testing, such as WES, is applied to rare disorders.
초록
배경
Coffin-Siris 증후군(Coffin-Siris syndrome, CSS)은 발달 지연, 지적 장애, 독특한 얼굴 특징, 다섯 번째 수족지의 저형성, 다모증 및 성긴 두발과 같은 특징으로 분류되는 매우 희귀한 질환이다. CSS는 현재 원인 유전자의 병원성 이형접합체 변이 및 결실/중복을 검출하는 분자유전학적 검사를 통해 진단된다.
방법
우리는 2017년 1월부터 2020년 12월까지 질병관리청 희귀 질환 진단 프로그램에 등록한 23명의 CSS 의심환자의 whole exome sequencing (WES) 보고서를 포함한 의무기록을 후향적으로 검토하였다. 통계 분석을 위해R 버전 4.2.0을 이용하여 Jaccard/Tanimoto 유사성 검정을 통한 군집 분석을 수행하였다.
결과
8건의 증례가 CSS로 진단되었다. ARID1B (NM_020732.3) Gln958*, Asn1320*, Gly1696* 및 Gly806Trpfs*, SMARCA4 (NM_001128849.1) Asn916Ser의 5가지 증례에서 새로운 변이가 확인되었다. 모든 ARID1B 증례에서 발작 등의 중추신경계 증상이 관찰되었고, 모든 SMARCA4 증례에서 다섯 번째 수족지의 저형성이 관찰되었다. SMARCA4 Asn916Ser은 de novo로 확인되었다. 유전형-표현형 분석에서 비교적 새로운 접근법인 군집 분석을 통해 증례들 간의 유사성 네트워크가 존재함을 확인하였다.
결론
우리는 CSS로 진단된 8명의 환자를 보고했으며 그 중 5명에서 ARID1B 또는 SMARCA4의 새로운 병원성 변이를 확인하였다. 본 연구는 유전자형-표현형 군집 분석 및 새로운 변이의 보고를 통해 CSS의 진단 및 분류 범주를 더욱 정교화하고, 향후 CSS에 대한 임상적 진단 기준을 명확히 하는 데 기여할 것으로 기대된다. 이 연구는 ARID1B 또는 SMARCA4에 의한 CSS의 표현형을 설명하는 데 기여한다. 이전에 잘 알려지지 않은 희귀 질환에 WES와 같은 보다 심층적인 유전자 검사가 적용됨에 따라 향후 연구에서는 전형적이지 않은 형질을 나타내는 환자에서 CSS를 더 용이하게 진단할 수 있을 것이다. 분자유전학적 검사를 통해 유전적 진단, 임상증상, 본 희귀질환의 분류 및 기준에 대한 추가 연구가 요구된다.
Coffin-Siris syndrome (CSS, MIM 135900) is a disorder characterized by distal phalanx, nail hypoplasia, or aplasia of the fifth finger or toe (digit), which may be accompanied by developmental or cognitive delay, characteristic facial features, hypotonia, hypertrichosis, microcephaly, and organ system abnormalities [1, 2]. The syndrome is caused by variants in several genes which are encoding components of Brahma-related gene 1 (BRG1)/ Brahma homologue (BRM)-Associated Factor (BAF) complex [3], which is one of the chromatin-remodeling complex families known in mammals [4, 5]. These genes include ARID1B, ARID1A, SMARCB1, SMARCA4, SMARCE1, ARID2, DPF2, SMARCC2, SOX11, SOX4, SMARCD1, and BICRA, each corresponding to CSS subtype 1-12, respectively [1, 2, 6]. Although phenotypic features for each CSS subtype have been established in previous studies [3], CSS cases without finger- or toe-related findings, which are representative phenotypes of some CSS subtypes, have also been reported recently [2].
A genotype-phenotype correlation study is an effective way to relate individual genetic backgrounds to specific diseases or characteristics, and is the basis of precision and genomic medicine [7]. The purpose of this study was to analyze genotype-phenotype correlations in patients with suspected developmental delay (DD) and intellectual disability (ID), and clinically suspected CSS, and to identify novel variants. This study was approved by the Institutional Review Board (IRB) of Seoul National University Hospital (IRB No. 2204-161-1320) under the Helsinki Declaration.
This retrospective study reviewed the electronic medical records of 23 suspected patients with CSS enrolled in a rare disease diagnostic program of the Korean Disease Control and Prevention Agency (KDCA) from January 2017 to December 2020. Patients were enrolled when they exhibited the following signs or symptoms, and demonstrated potential clinically suspected CSS: dysplasia of either the distal phalanx or the nail of the fifth digit, DD, ID, coarse facial features, hypertrichosis, seizures, and central nervous system (CNS) malformations. Among the 23 patients with suspected CSS, who requested for a rare disease diagnostic program, eight cases reported positive for CSS were included, and the remaining patients were excluded. For the CSS positive patients, basic information (reason for test, test date, diagnosis, age at diagnosis, sex), clinical manifestations (such as finger or toe hypoplasia, DD/ID, characteristic facial findings, hypotonia, scalp hair condition, cardiac anomaly, gastrointestinal tract anomaly, and CNS anomaly), and laboratory test results (reports from rare disease diagnostic program) were collected retrospectively from electronic medical records.
In the rare disease diagnosis program, whole-exome sequencing (WES), Sanger sequencing and multiplex ligation dependent probe amplification was performed. Then, variants were classified based on the American College of Medical Genetics and Genomics and the Association for Molecular Pathology guidelines [8].
The R version 4.2.0 (R Core Team, Vienna, Austria) was used for statistical analysis. The frequency and ratio of binary phenotypes per gene were calculated and visualized. A cluster analysis using Jaccard/Tanimoto similarity test [9] was performed to confirm if the cluster was formed according to genotype-phenotype, and the significance of the clusters. The Jaccard/Tanimoto coefficient is the ratio of union to intersection and is used to measure the similarity between binary data [9]. The network between clusters was expressed as a graph using the calculated P-value. The Leiden Open Variant Database [10], ClinVar, and literature review were used to determine if the pathogenic variants identified in this study have been previously reported.
The frequency of variant types and phenotypes for each causative gene is summarized in Table 1. In ARID1B, four cases of nonsense and each case of frameshift and splicing were identified, and two cases of missense were identified in SMARCA4 (Table 1). In the cases of ARID1B, CNS anomaly and DD/ID were observed in all cases, and hypertrichosis was observed in five cases. Among the CNS malformations, seizures were the most common in three cases, followed by shortening of corpus callosum in the two cases. In the cases of SMARCA4, hypoplasia of the fifth digit and unique facial features were observed in all cases (Table 1).
In Fig. 1A, the frequency of phenotypes observed by each gene is expressed as a bar plot. In the combination of cases, the number of phenotypes shared by the two cases was expressed as a matrix (Fig. 1B), and five phenotypes were observed identically in three cases of ARID1B. In a dendrogram based on the matrix (Fig. 1C), it was also confirmed that three of the ARID1B cases had the same phenotype completely, but some ARID1B cases showed more similar phenotypes to that of SMARCA4 cases. The adjacency matrix (Fig. 1D) was calculated using the Jaccard/Tanimoto similarity test of phenotype matrix (Fig. 1B), and case pairs showing significant similarity were indicated. A network with significant similarities between each case is graphically represented in Fig. 1E. The edge-connected nodes indicate a significant similarity. These nodes had common phenotypes: hypoplasia of the fifth digit, DD/ID, unique facial features, hypertrichosis, and CNS anomaly.
To the best of our knowledge, five novel variants were identified in this study (Table 2). In ARID1B cases constituting the genotype-phenotype cluster, three of the four cases showed novel variants (Gly1696*, Gly806Trpfs*, and Asn1320*). The novel variant (Asn916Ser) of SMARCA4 was a de novo variant identified through a trio test.
In this study, we confirmed that a subgroup of ARID1B has a significant similarity in terms of phenotype. In addition, to date, we have identified five new variants of CSS-causative genes including a de novo gene that was not previously reported to the best of our knowledge.
CSS is a genetically heterogeneous disease, and it has been previously established that the variants of ARID1B among the causative genes account for the largest proportion (up to 83%) of CSS [3, 11]. Although only two causative genes, ARID1B and SMARCA4, were identified in this study, the proportion of causative genes is congruous with previous studies [11]. Several ARID1B-associated CSS patients were previously reported to have various levels of DD/ID, hypertrichosis, and unique facial features such as low anterior hairline and thick eyebrows, however, hypoplasia of the fifth digit was observed in some ARID1B cases only [3, 11]. Patients with CSS having pathogenic variants of ARID1B are known to have relatively mild symptoms and normal growth [11]. ARID1B is one of the subunits that constitutes BAF complex, and is known to play a role in regulating the function of chromatin complexes [5]. In this epigenetic mechanism, ARID1B plays an important role in regulating gene activity by inducing the expression of proliferative genes, and promoting the cell cycle involved in neural stem cell differentiation [5, 12]. This could be inferred as the reason for CNS anomaly identified in all ARID1B cases in this study. The occurrence of episodes such as seizures in patients with suspected CSS, indicates the possibility of pathogenic variants in BAF-related genes, which should be confirmed [13]. In our study, the zygosity of every variant in ARID1B included heterozygous truncating variants, thus, it is expected to cause loss of function due to haploinsufficiency followed by the allelic decrease of functional gene product [2, 3, 5]. One splicing variant identified in ARID1B is located at the last base of exon 17, which is expected to cause alternative splicing in-silicon prediction using programs such as NNSPLICE, and GeneSplicer, and was reported to cause exon 17 skipping in a previous functional study [14]. In 40-80% of ARID1B-related diseases, including CSS, hypotonia is known to be present [15]. However, the medical records of ARID1B cases in this study did not contain explicit references to hypotonia. Hypotonia, which can be caused by anomalies in CNS or lower motor neurons, is a sign of decreased tone of the skeletal muscles and decreased resistance to passive stretching [16]. Seizures or global DD/ID including cognitive delay are frequently present in patients with central hypotonia, which has a mild peripheral muscular weakness [16, 17]. Several possibilities can be considered because hypotonia could not be identified in this study. In the case of CSS, hypotonia typically manifests in infancy [11], however, most patients in this study were diagnosed during their childhood or adulthood stage. Therefore, it is difficult to rule out the possibility that the evaluation of hypotonia was omitted during the medical history collection or the physical examination. However, considering most ARID1B cases in this study were also accompanied by delayed motor and cognitive development, and that DD was verified in four of the six cases, the likelihood of hypotonia cannot be fully ruled out. To unequivocally confirm this, a clinical re-evaluation of the patients with the ARID1B variants may be required.
SMARCA4 accounts for about 7% of CSS causes, which is the second-highest proportion [11]. In previous studies, it was established that patients with CSS with SMARCA4 variants almost always have hypoplasia of the fifth digit, many of them have hypotonia and behavioral abnormalities, and half of them have DD/ID [3, 11]. SMARCA4 is also a component of the BAF-complex, which forms a central ATPase. Heterozygous missense variants in SMARCA4A have a dominant-negative effect rather than haploinsufficiency, altering chromatin accessibility and making it difficult to bind to the super-enhancer [2, 18]. All our SMARCA4 cases were identified as heterozygous missense, all of them showed hypoplasia of the fifth digit, and half of them showed DD and hypotonia. These are consistent with the results from previous research [2, 11, 18]. In the case of a confirmed de novo variant of SMARCA4 Asn-916Ser, the patient’s older brother, sister, and parents were not affected, and it was confirmed that they did not have the variant in the trio test. The following symptoms were identified in this particular patient: a dysmorphic face, dysplasia of corpus callosum, dysmorphism of the second toe, and patent ductus arteriosus. Short stature, generalized laxity and hypotonia were presented as musculoskeletal manifestations. Generally, de novo variants are more harmful than other genetic variants inherited because it has been subjected to relatively less stringent evolutionary selection [19]. De novo variants are known to be a major cause of early onset genetic disorders such as Kabuki and CSS [19]. De novo variants in each BAF-related genes (ARID1A, ARID1B, SMARCB1, SMARCA4, SMARCA2, and SMARCE1) were reported to cause CSS, suggesting that abnormalities of BAF-complex cause abnormal neurodevelopment of CSS [20]. In addition, the variant is located in the ATP-ase domain of SMARCA4, and the site is also a mutational hotspot [21]. Missense variants in the SMARCA4 ATPase domain impair the regulatory mechanism of PRC1 protein related to methylation, thereby reducing the chromatin remodeling activity of the BAF-complex [21, 22].
There are some caveats in interpreting the results of this study. First, there may be limitations in generalization because the sample size is too small and patients were tested according to their clinical manifestation; one alternative solution is to analyze multi-omics together with genomic information. Second, a cluster with similar phenotypic networks was identified in a subset of specific causative genes, but owing to the nature of the retrospective study, CSS phenotypes that are not mentioned in the electronic medical record may exist, which may lead to changes in phenotypic networks and consequently, new clusters may emerge or existing clusters may disappear. Thus, it is expected that the cases that formed a single-member cluster in this study could be reclassified according to additional phenotypic information.
In conclusion, we reported four patients with a novel ARID1B variant and one patient with a novel SMARCA4 variant which is de novo. We also identified that there was a significant genotype-phenotype correlation between cases. This study contributes to describing the CSS phenotype caused by ARID1B or SMARCA4 variant. Future studies may facilitate easier diagnosis of CSS in patients who present with atypical traits using more in-depth genetic testing, such as WES, when applied to previously poorly known rare disorders. Molecular genetic testing may lead to further research on the genetic diagnosis, clinical symptoms, classification of this rare disease and criteria.
REFERENCES
1. Vasko A, Drivas TG, Schrier Vergano SA. 2021; Genotype-Phenotype Correlations in 208 Individuals with Coffin-Siris Syndrome. Genes. 12:937. DOI: 10.3390/genes12060937. PMID: 34205270. PMCID: PMC8233770.
2. Liu M, Wan L, Wang C, Yuan H, Peng Y, Wan N, et al. 2022; Coffin-Siris syndrome in two chinese patients with novel pathogenic variants of ARID1A and SMARCA4. Genes Genomics. 44:1061–70. DOI: 10.1007/s13258-022-01231-2. PMID: 35353340.
3. Kosho T, Miyake N, Carey JC. 2014; Coffin-Siris syndrome and related disorders involving components of the BAF (mSWI/SNF) complex: Historical review and recent advances using next generation sequencing. Am J Med Genet C Semin Med Genet. 166C:241–51. DOI: 10.1002/ajmg.c.31415. PMID: 25169878.
4. Alfert A, Moreno N, Kerl K. 2019; The BAF complex in development and disease. Epigenetics Chromatin. 12:19. DOI: 10.1186/s13072-019-0264-y. PMID: 30898143. PMCID: PMC6427853.
5. Sim JC, White SM, Lockhart PJ. 2015; ARID1B-mediated disorders: Mutations and possible mechanisms. Intractable Rare Dis Res. 4:17–23. DOI: 10.5582/irdr.2014.01021. PMID: 25674384. PMCID: PMC4322591.
6. OMIM®, # 135900 COFFIN-SIRIS SYNDROME 1; CSS1. https://www.omim.org/entry/135900. Update on Feb 2020.
7. Guo X, Song Y, Liu S, Gao M, Qi Y, Shang X. 2021; Linking genotype to phenotype in multi-omics data of small sample. BMC Genomics. 22:537. DOI: 10.1186/s12864-021-07867-w. PMID: 34256701. PMCID: PMC8278664.
8. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. 2015; Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–24. DOI: 10.1038/gim.2015.30. PMID: 25741868. PMCID: PMC4544753.
9. Chung NC, Miasojedow B, Startek M, Gambin A. 2019; Jaccard/Tanimoto similarity test and estimation methods for biological presence-absence data. BMC Bioinformatics. 20(Suppl 15):644. DOI: 10.1186/s12859-019-3118-5. PMID: 31874610. PMCID: PMC6929325.
10. Fokkema IFAC, Kroon M, López Hernández JA, Asscheman D, Lugtenburg I, Hoogenboom J, et al. 2021; The LOVD3 platform: efficient genome-wide sharing of genetic variants. Eur J Hum Genet. 29:1796–803. DOI: 10.1038/s41431-021-00959-x. PMID: 34521998. PMCID: PMC8632977.
11. Schrier Vergano S, Santen G, Wieczorek D, Wollnik B, Matsumoto N, Deardorff MA. Coffin-Siris Syndrome. https://www.ncbi.nlm.nih.gov/books/NBK131811/. Update on Aug 2021.
12. Moffat JJ, Smith AL, Jung EM, Ka M, Kim WY. 2022; Neurobiology of ARID1B haploinsufficiency related to neurodevelopmental and psychiatric disorders. Mol Psychiatry. 27:476–89. DOI: 10.1038/s41380-021-01060-x. PMID: 33686214. PMCID: PMC8423853.
13. Santen GW, Aten E, Vulto-van Silfhout AT, Pottinger C, van Bon BW, van Minderhout IJ, et al. 2013; Coffin-Siris syndrome and the BAF complex: genotype-phenotype study in 63 patients. Hum Mutat. 34:1519–28. DOI: 10.1002/humu.22394. PMID: 23929686.
14. Hoyer J, Ekici AB, Endele S, Popp B, Zweier C, Wiesener A, et al. 2012; Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability. Am J Hum Genet. 90:565–72. DOI: 10.1016/j.ajhg.2012.02.007. PMID: 22405089. PMCID: PMC3309205.
15. Vergano SA, van der Sluijs PJ, Santen G. ARID1B-Related Disorder. https://www.ncbi.nlm.nih.gov/books/NBK541502/. Updated on May 2019.
16. Lisi EC, Cohn RD. 2011; Genetic evaluation of the pediatric patient with hypotonia: perspective from a hypotonia specialty clinic and review of the literature. Dev Med Child Neurol. 53:586–99. DOI: 10.1111/j.1469-8749.2011.03918.x. PMID: 21418198.
17. Harris SR. 2008; Congenital hypotonia: clinical and developmental assessment. Dev Med Child Neurol. 50:889–92. DOI: 10.1111/j.1469-8749.2008.03097.x. PMID: 19046184.
18. Hodges HC, Stanton BZ, Cermakova K, Chang CY, Miller EL, Kirkland JG, et al. 2018; Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat Struct Mol Biol. 25:61–72. DOI: 10.1038/s41594-017-0007-3. PMID: 29323272. PMCID: PMC5909405.
19. Veltman JA, Brunner HG. 2012; De novo mutations in human genetic disease. Nat Rev Genet. 13:565–75. DOI: 10.1038/nrg3241. PMID: 22805709.
20. Tsurusaki Y, Okamoto N, Ohashi H, Kosho T, Imai Y, Hibi-Ko Y, et al. 2012; Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 44:376–8. DOI: 10.1038/ng.2219. PMID: 22426308.
21. Fernando TM, Piskol R, Bainer R, Sokol ES, Trabucco SE, Zhang Q, et al. 2020; Functional characterization of SMARCA4 variants identified by targeted exome-sequencing of 131,668 cancer patients. Nat Commun. 11:5551. DOI: 10.1038/s41467-020-19402-8. PMID: 33144586. PMCID: PMC7609548.
22. Stanton BZ, Hodges C, Calarco JP, Braun SM, Ku WL, Kadoch C, et al. 2017; Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet. 49:282–8. DOI: 10.1038/ng.3735. PMID: 27941795. PMCID: PMC5373480.
Fig. 1
Frequency of phenotypes observed using gene and cluster analysis. (A) The frequency of phenotypes observed in each causative gene was expressed as a percentage of the total cases of each gene. (B) The number of same phenotypes observed in the combination of cases was expressed as a matrix. (C) A dendrogram based on the matrix depicted in (B). (D) Cluster analysis using Jaccard/Tanimoto similarity test for (B). (E) Networks with significant phenotypic similarities between each case was plotted as a graph based on the matrix depicted in (D), when the P≤0.01 was applied to (D) as a cutoff. The distance between each node is meaningless.
Abbreviations: DD/ID, developmental delay/intellectual disability; GI, gastrointestinal; GU, genitourinary; CNS, central nervous system.
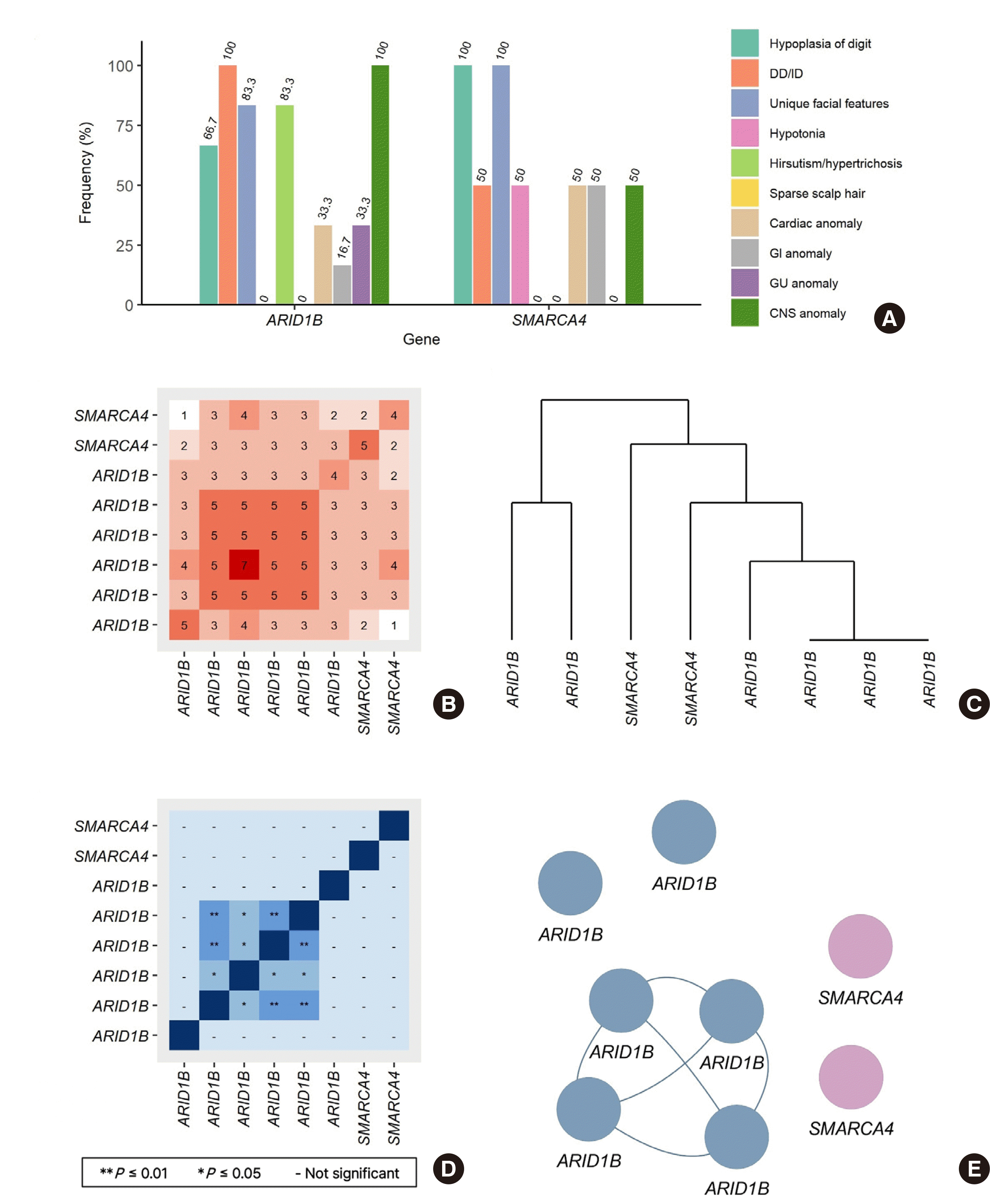
Table 1
Types of variants and phenotypes by causative gene in study population
Table 2
Genetic variants and ACMG classification identified in cases and its novelty status




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download