Abstract
OPHN1 is located in Xq12 and acts as a regulator for the development of neural tissues in a fetus. Various spectrums of X-linked intellectual disabilities (XLIDs) can occur due to a loss of function in the OPHN1 gene. In this case study, the authors report two cases of OPHN1-related XLID found in one family in Korea. A 7-year-old boy presented with speech development delay, intellectual disability, and an epilepsy event. There were no specific perinatal history or test results except suggestive mega cisterna magna. His younger brother had a similar phenotype. A chromosomal microarray (CMA) test showed a hemizygous 414 kbp deletion (chrX: 67,582,399-67,997,055) in both brothers, and thereafter a deletion of exons 1 and 2 of OPHN1 was confirmed via PCR. In summary, it is difficult to specify the causative gene of an intellectual disability by symptoms alone. Therefore, CMA can be used as an important diagnostic test along with tests such as whole exome sequencing.
초록
OPHN1 유전자는 Xq12에 위치하며 그 산물인 oligophrenin-1은 태아에서 신경 조직의 발달을 조절하는 역할을 한다. 이러한 OPHN1 유전자의 기능 상실로 인해 다양한 양상의 X-연관 지적 장애(XLID)가 발생할 수 있다. 이에 저자들은 한국인 가족에서 발견된 OPHN1 관련 XLID 환자 두 명의 증례를 보고한다. 7세 소년이 언어 발달 지연, 지적 장애 및 간질을 주소로 내원하였으며, 거대대조(mega-cisterna magna) 소견을 제외하고는 특별한 주산기 병력이나 비정상 검사 결과는 없었다. 환아의 남동생도 비슷한 표현형을 보여 시행한 CMA (chromosomal microarray) 검사에서 두 형제 모두 이형접합체 414 kbp 결실(chrX: 67,582,399-67,997,055)이 관찰되었고, 이후 PCR 검사법를 이용해 OPHN1 엑손 1, 2의 결실을 확인하였다. 본 증례와 같이 증상만으로 지적 장애의 원인 유전자를 특정하기는 어려우며, 따라서 이러한 지적 장애 환자들에서 전장유전체분석과 더불어 CMA가 중요한 진단 검사법으로 활용될 수 있을 것으로 생각된다.
There are many genetic disorders or syndromes that cause intellectual disabilities, and they are usually accompanied by inconsistent symptoms or abnormalities. OPHN1, one of the causative genes of hereditary intellectual disability, is located in Xq12 and encodes an oligophrenin-1 protein that plays an important regulatory role in the development and migration of fetal nervous tissues [1]. As we are aware, from 1997 to the present, there have been several case reports of wide spectrums of X-linked intellectual disabilities (XLIDs) associated with variants of the OPHN1 gene. [2–9]. XLIDs caused by OPHN1 gene mutations have mental retardation and cerebellar hypoplasia in common, and other features such as epilepsy, ataxia, strabismus, urogenital abnormality, facial dysmorphism, and ventriculomegaly can also be observed. In this case study, we report two cases of OPHN1-associated XLID found in one family, the first to our knowledge, in Korea. The requirement for informed consent was waived (IRB No. 2022-05-060-001) by Institutional Review Board of Keimyung university Dongsan Hospital.
In 2015, a 23-month-old boy visited the rehabilitation department with a general developmental delay. He was his parent’s first child and was born at full term following a normal pregnancy via vaginal delivery; his body weight was 3,300 g. He presented with facial dysmorphisms, including a prominent forehead, ocular anomalies (bilateral ptosis, horizontal nystagmus, and impaired visual perception), a prominent nose with a wide nasal base, a high palate, dental cavities, and pes planus. A prominent cystic lesion in the retrocerebellar convexity suggesting mega cisterna magna findings was observed on the brain MRI; the laboratory tests showed no abnormal results with the 46,XY karyotyping. During his neurological examination when he was 7 years old, he exhibited poor fine motor coordination and the inability to organize activities of daily living. The psychological examination showed moderate intellectual disability with a low spatial orientation, a working memory with short-term memory capacity, emotional instability, and poor concentration. The personal and social adaptive functions were equivalent to that of a child aged 53 months (SQ=56.19). The receptive language age was 47 months and the expressive language age was 41 months.
In 2021, his younger brother, a 45-month-old male, visited the rehabilitation department with a developmental delay as well as a retro-cerebellar cystic mass and diffuse frontal cortical atrophy from birth. He was born at intrauterine pregnancy of 38 weeks with a body weight of 2,600 g. During the clinical examination, we observed the following characteristics: discrete facial dysmorphisms, pes planus, and functional scoliosis with poor posture. The psychomotor examination revealed delayed gross motor development with poor balance on walking ambulation and was equivalent to that of a child aged 30 months. The personal and social scales were equivalent to a child aged 30 months, and the language scale age was 25 months. He was also taking medicine for hypothyroidism.
A chromosomal microarray (CMA) test was performed with the GeneChip® System 3000Dx v.2 (Thermo Fisher Scientific, Waltham, MA, US) using a CytoScan™ Dx Assay Kit (Cat. No. 902420, Thermo Fisher Scientific), to confirm the hereditary intellectual developmental disorder. As a result of the CMA test, a hemizygous 360 kbp deletion (chrX: 67,636,890-67,997,055) containing a part of OPHN1, known as the causative gene for XLID, was discovered (Fig. 1A). To confirm the partial deletion of the OPHN1 gene, a PCR test was performed using primer pairs for exons 1, 2, 3, and 4, and the deletion of exons 1 and 2 was confirmed (Fig. 1B, C). The identical hemizygous deletion was found in the proband’s younger brother with similar symptoms. In additional CMA and PCR family tests, the heterozygous deletion was revealed in the unaffected mother and younger sister. The father showed no copy number variation in OPHN1. The primers and targets of the PCR study are described in Table 1.
Several types of mutations have been reported for XLIDs related to OPHN1 mutations, from single nucleotide substitutions to exon deletions, and they have different symptoms depending on the location and type of mutation [2]. For this reason, case reports including the exact variant description are important for genotype-phenotype relation studies. The OPHN1 exons 1 and 2 deletion observed in the cases in this study are suspected to have a similar effect to the case reported by Bienvenu et al. [3], which involved a female patient with mental retardation, epilepsy, ventriculomegaly, and strabismus. She had an X;12 balanced translocation and Billuart et al. [10] identified that this translocation happened in the second intron between exons 2 and 3 and revealed no expression of OPHN1 in the patient.
On the other hand, several studies on the physiological function and pathway related to the Rho-kinase activity of the OPHN1 gene have been reported [1, 11]. Recently, Wang et al. [12] investigated the exact mechanism of stress-related behavior commonly found in OPHN1 gene deficiency, and at the same time suggested its potential as a therapeutic target through animal model experiments. As such, in the case of hereditary intellectual disabilities, precise target treatments may appear depending on the results of future research, so it is important to identify the exact trigger gene for each patient and to accumulate accurate knowledge about the genotype-phenotype relationship. However, in many individual hereditary intellectual disabilities, it is difficult to differentiate between them based on their clinical symptoms alone due to inconsistent phenotypes. Therefore, when genetic intellectual disability is suspected due to the presence of family history as in these cases, it is expected that CMA can be used as an important diagnostic test along with other tests such as whole exome sequen cing.
REFERENCES
1. Barresi S, Tomaselli S, Athanasiadis A, Galeano F, Locatelli F, Bertini E, et al. 2014; Oligophrenin-1 (OPHN1), a gene involved in X-linked intellectual disability, undergoes RNA editing and alternative splicing during human brain development. PLoS One. 9:e91351. DOI: 10.1371/journal.pone.0091351. PMID: 24637888. PMCID: PMC3956665.
2. Schwartz TS, Wojcik MH, Pelletier RC, Edward HL, Picker JD, Holm IA, et al. 2019; Expanding the phenotypic spectrum associated with OPHN1 variants. Eur J Med Genet. 62:137–43. DOI: 10.1016/j.ejmg.2018.06.015. PMID: 29960046. PMCID: PMC6310103.
3. Bienvenu T, Der-Sarkissian H, Billuart P, Tissot M, Des Portes V, Brüls T, et al. 1997; Mapping of the X-breakpoint involved in a balanced X;12 translocation in a female with mild mental retardation. Eur J Hum Genet. 5:105–9. DOI: 10.1159/000484743. PMID: 9195162.

4. Bergmann C, Zerres K, Senderek J, Rudnik-Schöneborn S, Eggermann T, Häusler M, et al. 2003; Oligophrenin 1 (OPHN1) gene mutation causes syndromic X-linked mental retardation with epilepsy, rostral ventricular enlargement and cerebellar hypoplasia. Brain. 126:1537–44. DOI: 10.1093/brain/awg173. PMID: 12805098.
5. Santos-Rebouças CB, Belet S, Guedes de Almeida L, Ribeiro MG, Medina-Acosta E, Bahia PR, et al. 2014; A novel in-frame deletion affecting the BAR domain of OPHN1 in a family with intellectual disability and hippocampal alterations. Eur J Hum Genet. 22:644–51. DOI: 10.1038/ejhg.2013.216. PMID: 24105372. PMCID: PMC3992576.

6. Moortgat S, Lederer D, Deprez M, Buzatu M, Clapuyt P, Boulanger S, et al. 2018; Expanding the phenotypic spectrum associated with OPHN1 mutations: Report of 17 individuals with intellectual disability but no cerebellar hypoplasia. Eur J Med Genet. 61:442–50. DOI: 10.1016/j.ejmg.2018.03.002. PMID: 29510240.
7. Iida A, Takeshita E, Kosugi S, Kamatani Y, Momozawa Y, Kubo M, et al. 2018; A novel intragenic deletion in OPHN1 in a Japanese patient with Dandy-Walker malformation. Hum Genome Var. 6:1. DOI: 10.1038/s41439-018-0032-8. PMID: 30534410. PMCID: PMC6281661.
8. Bogliş A, Cosma AS, Tripon F, Bãnescu C. 2020; Exon 21 deletion in the OPHN1 gene in a family with syndromic X-linked intellectual disability: Case report. Medicine (Baltimore). 99:e21632. DOI: 10.1097/MD.0000000000021632. PMID: 32872024. PMCID: PMC7437857.
9. Nuovo S, Brankovic V, Caputi C, Casella A, Nigro V, Leuzzi V, et al. 2021; Novel unconventional variants expand the allelic spectrum of OPHN1 gene. Am J Med Genet A. 185:1575–81. DOI: 10.1002/ajmg.a.62144. PMID: 33638601.
10. Billuart P, Bienvenu T, Ronce N, des Portes V, Vinet MC, Zemni R, et al. 1998; Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 392:923–6. DOI: 10.1038/31940. PMID: 9582072.
11. Nadif Kasri N, Nakano-Kobayashi A, Van Aelst L. 2011; Rapid synthesis of the X-linked mental retardation protein OPHN1 mediates mGluR-dependent LTD through interaction with the endocytic machinery. Neuron. 72:300–15. DOI: 10.1016/j.neuron.2011.09.001. PMID: 22017989. PMCID: PMC3206629.
12. Wang M, Gallo NB, Tai Y, Li B, Van Aelst L. 2021; Oligophrenin-1 moderates behavioral responses to stress by regulating parvalbumin interneuron activity in the medial prefrontal cortex. Neuron. 109:1636–56.e8. DOI: 10.1016/j.neuron.2021.03.016. PMID: 33831348. PMCID: PMC8141044.
Fig. 1
The results of the CMA and PCR tests of OPHN1 exons 1–4. (A) The weighted log2 ratio and copy number state of the proband (P), his brother (B), sister (S), father (F), and mother (M). The proband and brother showed a hemizygous 414 kbp deletion (chrX: 67,582,399-67,997,055). His mother and sister had a heterozygous deletion in the same region. The father showed no abnormal findings. (B) Gel electrophoresis of the PCR products amplifying exons 1–2 of OPHN1 with the sample of the proband and family members supported the CMA findings. The proband and brother showed a complete loss of OPHN1 exons 1–2, while the mother and sister showed exons thought to be derived from conserved alleles. Primers for exons 10 of G6PD (154 bp on the band below) were used as the internal control in all the PCR tests, and ladders ranging in size from 100 to 1,300 bp were shown in the center. (C) Gel electrophoresis of the PCR products amplifying OPHN1 exons 3–4 with the sample of the proband and family members supported the CMA findings. Exons 3–4 were detected in all the family members.
Abbreviation: CMA, chromosomal microarray.
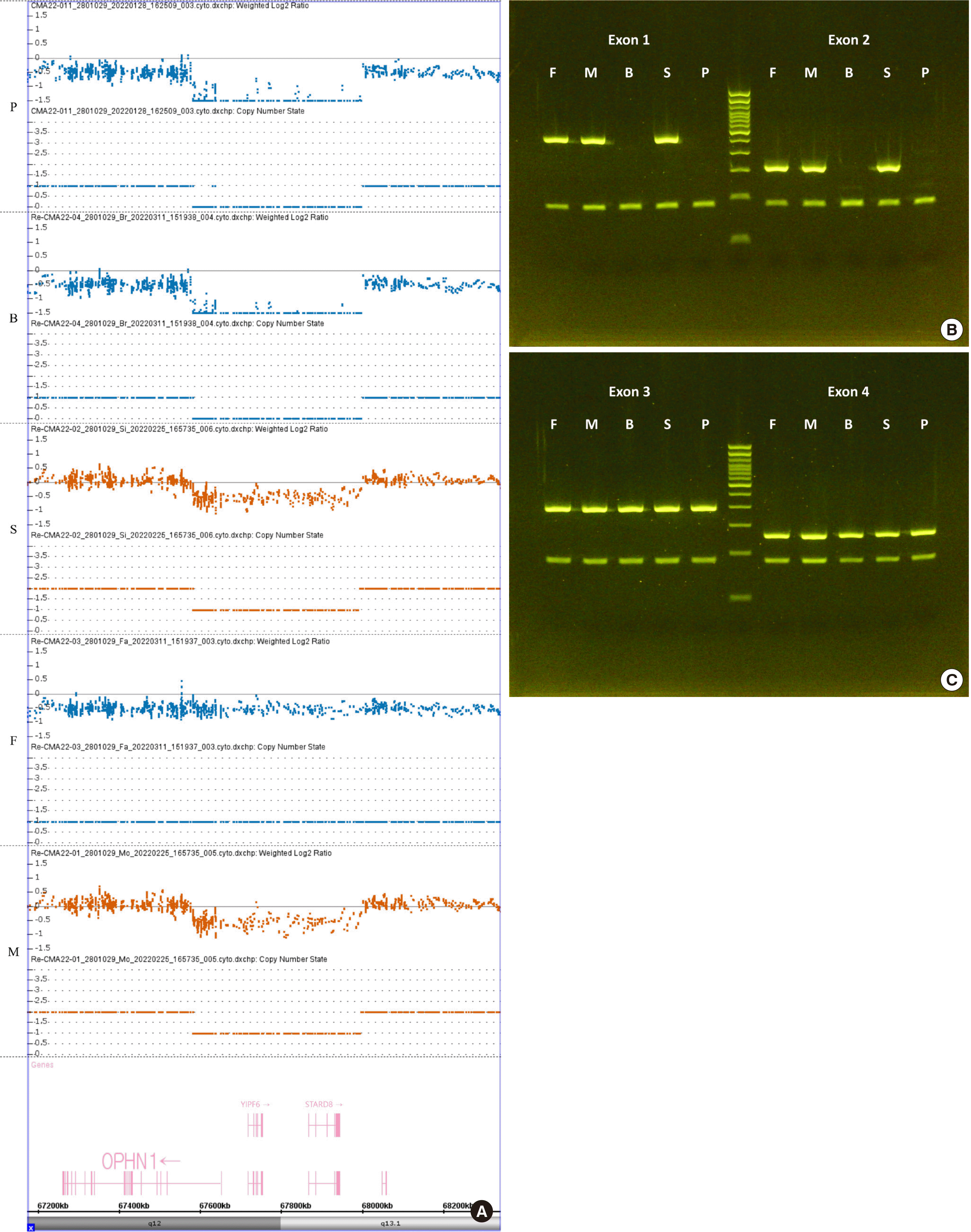
Table 1
Description of the primers used for the PCR test of OPHN1 exons 1–4




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download