Abstract
A 77-year-old man with pain in his left hand after being pricked by a beanstalk visited an emergency center. Symptoms and laboratory findings suggested infectious tenosynovitis. Using the Bruker Biotyper (Bruker Daltonics, Germany), Kosakonia cowanii was identified in the swab culture and was confirmed by full-length 16S ribosomal RNA (rRNA) and gyrB sequencing. To the best of our knowledge, this is the first reported case of K. cowanii infection in Korea. Performing only 16S rRNA sequencing was insufficient for accurate identification, and gyrB sequencing or using the Bruker Biotyper (Bruker Daltonics, Germany) confirmed the identification. A positive sorbitol fermentation test can indicate the microorganism to be K. cowanii even if the VITEK 2 system (bioMérieux, France) identifies it as Pantoea spp. Additionally, K. cowanii may be resistant to beta-lactam antibiotics, including cefazolin; therefore, it may be appropriate to use carbapenems, third- or fourth-generation cephalosporins, aminoglycosides, fluoroquinolones, and trimethoprim/sulfamethoxazole. Finally, antibiotic susceptibility testing (AST) should be conducted using a reference method, and Etest AST is unreliable for K. cowanii, especially with beta-lactam/beta-lactamase inhibitors.
초록
77세 남성이 콩줄기에 찔려 생긴 왼손 통증으로 응급센터를 방문하였다. 전형적인 증상과 실험실 소견은 감염성 건초염을 강하게 시사했다. 면봉 배양에서 Kosakonia cowanii가 Bruker Biotyper (Bruker Daltonics, Germany)로 확인되었으며, 전체 16S 리보솜 RNA와 gryB 염기서열분석으로 확인되었다. 이는 국내 최초로 보고된 K. cowanii 인체 감염 사례이다. 16S rRNA 염기서열분석만 시행하는 것은 정확한 동정을 하기에 충분하지 않았으며, gyrB 염기서열분석을 추가하거나 Bruker Biotyper (Bruker Daltonics)를 사용하면 동정이 가능하다. 혹은 만약 Pantoea spp.가 Vitek 2 system (bioMérieux, France)에서 의심되지만 소르비톨 발효가 양성인 경우 K. cowanii를 의심할 수 있다. 또한 K. cowanii는 세파졸린을 포함한 베타락탐 항생제에 내성이 있을 수 있으므로 그러한 경우에는 카바페넴, 3세대 또는 4세대 세팔로스포린, 아미노글리코사이드, 플루오로퀴놀론 또는 트리메토프림/술파메톡사졸을 사용하는 것이 적절할 수 있다. 마지막으로 항생제감수성검사는 표준검사 방법을 사용해야 하며 Etest를 이용한 항생제감수성검사는 아직 신뢰하기 어렵다.
A 77-year-old male, with underlying coronary artery obstructive disease and diseases of idiopathic pulmonary fibrosis, hypertension, diabetes, and dyslipidemia, visited the emergency center presenting with left hand pain. He was administered aspirin, trim-etazidine, sacubitril/valsartan, torasemide, neustatin, metformin, dapagliflozin, lansoprazole, theobromine, bromhexine, and lafutidine at our hospital. Three days prior to visiting the emergency center, he fell and his left palm was pricked by a beanstalk. A day later, he visited a local hospital for treatment and was administered cefazolin intravenously. Computed tomography (CT) was performed at a local hospital and infectious tenosynovitis of the left hand was suspected. He was then referred to our hospital for emergency surgery. Approximately 2 cm of linear abrasion with pus and sanguineous discharge were observed in the middle of the left palm (Fig. 1). The left hand was diffusely swollen, and the patient complained of heating sensation, tenderness, and pain. No neurological deficits or blood circulation problems were found. Vital signs were 103/69 mmHg for systolic/diastolic blood pressure, 118/min for pulse rate, 18/min for respiratory rate, and 36.8°C for body temperature. The routine laboratory test detected leukocyte count of 10,820/mm3 with 72.7% of neutrophils, erythrocyte sedimentation rate of 26 mm/h (reference interval [RI], 0.0–22.0 mm/h), C-reactive protein of 119.4 mg/L (RI, 0–8 mg/L), and procalcitonin of 0.14 ng/mL (RI, 0.00–0.50 ng/mL). Typical symptoms and laboratory findings strongly suggested infectious tenosynovitis. The patient was referred to another hospital because of emergency surgery difficulties after collecting the wound discharge for microbiological investigations.
The white blood cells (WBCs) were 3+ in gram staining of wound discharge, but no organism was seen by microscopy. However, few medium-sized, colorless, smooth, convex, and glistening colonies were observed on blood agar and few medium-sized, pink, smooth, convex, punctate, umbilicated, and glistening colonies were observed on MacConkey agar plates incubated at 35°C under 5% CO2 for 24 hours (Fig. 2). The microorganisms were medium-to-long, plump, gram-negative rods (Fig. 3) and were oxidase-negative and catalase-positive. In the triple sugar iron medium, the slant and the butt were yellow, and gas generation was confirmed, but hydrogen sulfide was absent. The Bruker Biotyper (Bruker Daltonics, Bremen, Germany) identified the microorganism as Kosakonia cowanii with a score of 2.12, which is reliable for species level identification according to the manufacturer’s instruction. As K. cowanii is a rarely identified bacteria in clinical laboratories, the VITEK 2 system (bioMérieux, Marcy l’Étoile, France) and VITEK MS (bioMérieux) were additionally used for confirming the results of bacterial identification. However, the former was unable to identify this isolate and the latter misidentified the isolate as Pantoea spp. with 98% probability (Table 1). To solve the discrepancy, full-length 16S ribosomal RNA (rRNA) gene sequencing was performed for species-level identification, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [1]. The amplicon was sequenced by Macrogen (Korea) to yield 16S rRNA gene sequence. The length of the sequence was 1455 bp. BLAST (http://www.ncbi.nlm.nih.gov/) search was conducted and the EzBioCloud (http://eztaxon-e.ezbiocloud.net/) [2] was employed using the sequence. For species level, we considered over 99% homology as acceptable criteria. However, we could not identify the sequence accurately because some of the sequences with over 99% homology included the 16S rRNA gene sequence of K. cowanii, Salmonella bongori, and Atlantibacter hermannii. Based on a previous report, we additionally performed gyrB sequencing using the primers UP1 and 181r [3]. Finally, the microorganism was identified as K. cowanii with 99.04% identity, based on similarity with 521 base pairs of K. cowanii strain Pa82 (GenBank accession number CP069319.1).
The antimicrobial susceptibility test (AST) was performed using MicroScan (Beckman Coulter, CA, USA) (Table 2). Antimicrobial susceptibility was interpreted according to the CLSI M100 guidelines [4]. This isolate was resistant to ampicillin, ampicillin/sulbactam, amoxicillin/clavulanic acid, cefoxitin, and cefuroxime. It was susceptible to penicillin in a dose dependent manner, intermediately susceptible to colistin, and highly susceptible to the other tested antibiotics. Due to the concentration range of antibiotics, the minimal inhibitory concentrations (MIC) of ciprofloxacin and levofloxacin could not be interpreted precisely by MicroScan. We also performed Etest (bioMérieux) (Table 2). The Etest results were concordant with the MicroScan results, except in the case of levofloxacin and amoxicillin/clavulanic acid. The levofloxacin MIC in the Etest was interpreted as susceptible. Interestingly, the MIC of amoxicillin/clavulanic acid was significantly different, resulting in a “resistant” result according to MicroScan and a “susceptible” result according to Etest.
The name Enterobacter cowanii was first proposed in 2000 and changed to K. cowanii in 2013 [5, 6]. The Kosakonia spp. was differentiated from the Enterobacter spp. using the multilocus sequence analysis method, which analyzed the genes of gyrB, rpoB, infB, and atpD [6]. There are nine species of Kosakonia, including K. cowanii. K. cowanii belongs to the family Enterobacteriaceae, order Enterobacterales, and class Gammaproteobacteria. It is a gram-negative rod, facultatively anaerobic, oxidase-negative, catalase-positive, and non-spore-forming bacterium, which ferments glucose and reduces nitrates to nitrites [7]. Most K. cowanii individuals are motile and have peritrichous flagella; they grow well on MacConkey agar and have pink, smooth, convex, punctate, umbilicated, glistening colonies [7].
K. cowanii is reported to cause bacterial wilt in some plants [8, 9]; however, its human pathogenicity is not well known. Few case reports have described K. cowanii infection, including bacteremia in neonates [3], rhabdomyolysis as a result of bacteremia preceded by a rose thorn prick [10], and acute cholecystitis [11]. K. cowanii infection seems to occur from exogenous sources [10] and endogenous sources, such as the patient’s microbiota [3, 11]. The possibility of K. cowanii infection must be considered in patients with plant-related injuries. In this case, the infectious synovitis was caused by the prick of a beanstalk.
K. cowanii grows well in routine laboratory cultures; however, its identification method is limited. The VITEK MS (bioMérieux) and VITEK 2 systems (bioMérieux) provide no information on K. cowanii and the VITEK 2 system (bioMérieux) misidentified it as Pantoea spp., which was the same as that obtained in a previous report [3, 11]. Additionally, the colony morphology is very similar between the Pantoea spp., and K. cowanii, making it difficult to differentiate the two organisms [7, 12]. The biochemical characteristics of K. cowanii are similar to those of Pantoea spp., but if the organisms produce gases during glucose, dulcitol or sorbitol fermentation, they are more likely to be K. cowanii [5]. In this case, K. cowanii showed the same biochemical characteristics as previously reported (Table 1) [5, 7], and importantly, with positive sorbitol fermentation. For Pantoea agglomerans ATCC 27155 and Pantoea dispersa ATCC 14589, sorbitol fermentation is negative [13], although there are reports of some Pantoea spp. isolates showing sorbitol fermentation [14, 15]. In addition, full-length 16S rRNA sequencing is insufficient to differentiate among K. cowanii, S. bongori, and A. hermannii; therefore, 16S rRNA with additional gyrB sequencing is the most accurate method to identify K. cowanii. The Bruker Biotyper (Bruker Daltonics) with reliable scores is also conveniently available in clinical laboratories. Additionally, a positive sorbitol fermentation test can indicate the microorganism to be K. cowanii even if the VITEK 2 system (bioMérieux) identifies it as Pantoea spp. in patients who are pricked by plants or have a plant-related job.
The antibiograms of the isolates in this study and previous reports are shown in Table 2 [3, 10, 11]. The Kosakonia genomes were known to lack AmpC β-lactamase [16]; however, in this study, the isolate seemed to have a mechanism for acquired resistance. The isolate was resistant to first- and second-generation cephalosporin antibiotics; therefore, it was likely to be resistant to cefazolin. In general, cefazolin is widely used for skin and soft tissue infections; however, it may not be appropriate in this case. It may be appropriate to use carbapenems, third- or fourth-generation cephalosporins, aminoglycosides, fluoroquinolones, or trimetho-prim/sulfamethoxazole. We also performed some antibiotic susceptibility tests using Etest, most of which showed concordant results with MicroScan. However, the AST results for amoxicillin/clavulanic acid showed discrepancies. Some reports about the discrepancy between the broth microdilution method and other beta-lactams/beta-lactamase inhibitors for Enterobacteriaceae exist [17-19], which may be because beta-lactamase is better expressed in liquid media. Therefore, Etest for K. cowanii should be avoided, especially with beta-lactam/beta-lactamase inhibitors. Meanwhile, Pantoea spp. is also usually susceptible to many antibiotics, but resistance to amoxicillin/clavulanic acid or ampicillin appears to be common [20].
The limitation of this study was that we could not follow-up on the patient’s prognosis. In addition, we did not perform cefazolin AST. Therefore, we could not predict or evaluate the response of this isolate to cefazolin treatment. Finally, whole-genome sequencing was not performed. This might be helpful in determining the antimicrobial resistance mechanisms of K. cowanii.
In conclusion, K. cowanii can cause infectious tenosynovitis, and this is the first case of human infection in Korea. 16S rRNA sequencing is not enough to identify K. cowanii, and gyrB sequencing should be performed as well. Bruker Biotyper (Bruker Daltonics) also seems to be good for identify it. A positive sorbitol fermentation test can indicate the microorganism to be K. cowanii even if the VITEK 2 system (bioMérieux) detects the presence of Pantoea spp. Additionally, K. cowanii may be resistant to beta-lactam antibiotics, including cefazolin; therefore, it may be appropriate to use carbapenems, third- or fourth-generation cephalosporins, aminoglycosides, fluoroquinolones, and trimethoprim/sulfamethoxazole. Furthermore, AST should be conducted using a reference method, and AST using the Etest is unreliable for K. cowanii, especially with beta-lactam/beta-lactamase inhibitors.
REFERENCES
1. Clinical, Laboratory Standards Institute. 2018. Interpretive criteria for identification of bacteria and fungi by targeted DNA sequencing. 2nd ed. CLSI guideline MM18. Clinical and Laboratory Standards Institute;Wayne, PA:
2. Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, et al. 2017; Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 67:1613–7. DOI: 10.1099/ijsem.0.001755. PMID: 28005526. PMCID: PMC5563544.
3. Duployez C, Edun-Renard ME, Kipnis E, Dessein R, Le Guern R. 2021; Bacteremia due to Kosakonia cowanii in a preterm neonate. J Pediatr Infect Dis. 16:183–6. DOI: 10.1055/s-0040-1721448.
4. Clinical, Laboratory Standards Institute. 2022. Performance standards for antimicrobial susceptibility testing. 32nd ed. CLSI supplement M100. Clinical and Laboratory Standards Institute;Wayne PA:
5. Inoue K, Sugiyama K, Kosako Y, Sakazaki R, Yamai S. 2000; Enterobacter cowaniisp. nov., a new species of the family Enterobacteriaceae. Curr Microbiol. 41:417–20. DOI: 10.1007/s002840010160. PMID: 11080391.
6. Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P. 2013; Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol. 36:309–19. DOI: 10.1016/j.syapm.2013.03.005. PMID: 23632228.
7. Mardaneh J, Soltan-Dallal MM. 2014; Isolation and identification of E. cowanii from powdered infant formula in NICU and determination of antimicrobial susceptibility of isolates. Iran J Pediatr. 24:261–6.
8. Zhang Y, Wang B, Li Q, Huang D, Zhang Y, Li G, et al. 2022; Isolation and Complete Genome Sequence Analysis of Kosakonia cowanii Pa82, a Novel Pathogen Causing Bacterial Wilt on Patchouli. Front Microbiol. 12:818228. DOI: 10.3389/fmicb.2021.818228. PMID: 35095821. PMCID: PMC8795763.
9. Sarkar S, Chaudhuri S. 2015; New report of additional enterobacterial species causing wilt in West Bengal, India. Can J Microbiol. 61:477–86. DOI: 10.1139/cjm-2015-0017. PMID: 26040797.
10. Washio K, Yamamoto G, Ikemachi M, Fujii S, Ohnuma K, Masaki T. 2018; Rhabdomyolysis due to bacteremia from Enterobacter cowanii caused by a rose thorn prick. J Dermatol. 45:e313–4. DOI: 10.1111/1346-8138.14341. PMID: 29696688.
11. Berinson B, Bellon E, Christner M, Both A, Aepfelbacher M, Rohde H. 2020; Identification of Kosakonia cowanii as a rare cause of acute cholecystitis: case report and review of the literature. BMC Infect Dis. 20:366. DOI: 10.1186/s12879-020-05084-6. PMID: 32448208. PMCID: PMC7245821.
12. Mardaneh J, Dallal MM. 2013; Isolation, identification and antimicrobial susceptibility of Pantoea (Enterobacter) agglomerans isolated from consumed powdered infant formula milk (PIF) in NICU ward: First report from Iran. Iran J Microbiol. 5:263–7.
13. Gavini F, Mergaert J, Beji A, Mielcarek C, Izard D, Kersters K, et al. 1989; Transfer of Enterobacter agglomerans (Beijerinck 1888) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and Description of Pantoea dispersa sp. nov. Int J Syst Evol Microbiol. 39:337–45. DOI: 10.1099/00207713-39-3-337.
14. Rezzonico F, Smits TH, Montesinos E, Frey JE, Duffy B. 2009; Genotypic comparison of Pantoea agglomerans plant and clinical strains. BMC Microbiol. 9:204. DOI: 10.1186/1471-2180-9-204. PMID: 19772624. PMCID: PMC2764716.
15. Farmer JJ 3rd, Davis BR, Hickman-Brenner FW, McWhorter A, Huntley-Carter GP, Asbury MA, et al. 1985; Biochemical identification of new species and biogroups of Enterobacteriaceae isolated from clinical specimens. J Clin Microbiol. 21:46–76. DOI: 10.1128/jcm.21.1.46-76.1985. PMID: 3881471. PMCID: PMC271578.

16. Bhatti MD, Kalia A, Sahasrabhojane P, Kim J, Greenberg DE, Shelburne SA. 2017; Identification and whole genome sequencing of the first case of Kosakonia radicincitans causing a human bloodstream infection. Front Microbiol. 8:62. DOI: 10.3389/fmicb.2017.00062. PMID: 28174569. PMCID: PMC5258702.
17. Soares A, Pestel-Caron M, de Rohello F, Bourgoin G, Boyer S, Caron F. 2020; Area of technical uncertainty for susceptibility testing of amoxicillin/clavulanate against Escherichia coli: Analysis of automated system, Etest and disk diffusion methods compared to the broth microdilution reference. Clin Microbiol Infect. 26:1685. e1-6. DOI: 10.1016/j.cmi.2020.02.038. PMID: 32151599.
18. Giani T, Morosini MI, D'Andrea MM, García-Castillo M, Rossolini GM, Cantón R. 2012; Assessment of the Phoenix™ automated system and EUCAST breakpoints for antimicrobial susceptibility testing against isolates expressing clinically relevant resistance mechanisms. Clin Microbiol Infect. 18:E452–8. DOI: 10.1111/j.1469-0691.2012.03980.x. PMID: 22909279.
19. Donay JL, Mathieu D, Fernandes P, Prégermain C, Bruel P, Wargnier A, et al. 2004; Evaluation of the automated Phoenix system for potential routine use in the clinical microbiology laboratory. J Clin Microbiol. 42:1542–6. DOI: 10.1128/JCM.42.4.1542-1546.2004. PMID: 15071001. PMCID: PMC387561.
20. Gajdács M. 2019; Epidemiology and antibiotic resistance trends of Pantoea species in a tertiary-care teaching hospital: A 12-year retrospective study. Develop Health Sci. 2:72–5. DOI: 10.1556/2066.2.2019.009.
Fig. 1
Clinical manifestations of the patient. Approximately 2 cm of linear abrasion with pus and sanguineous discharge were observed in the middle of the left palm. The patient’s left hand was diffusely swollen.
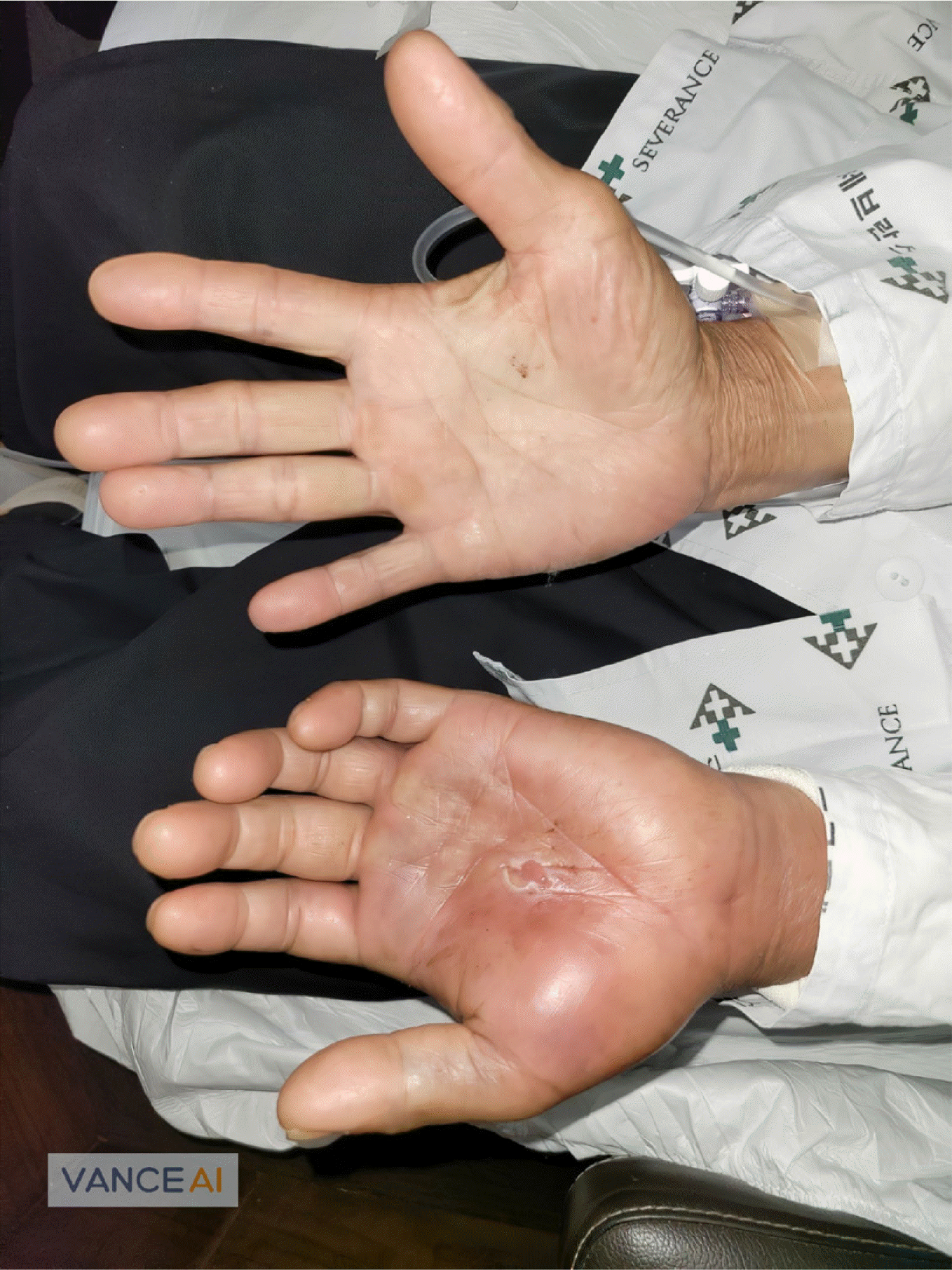
Fig. 3
Gram-stain of Kosakonia cowanii (×1,000). Medium-to-long, plump, gram-negative rods were observed by Gram staining.
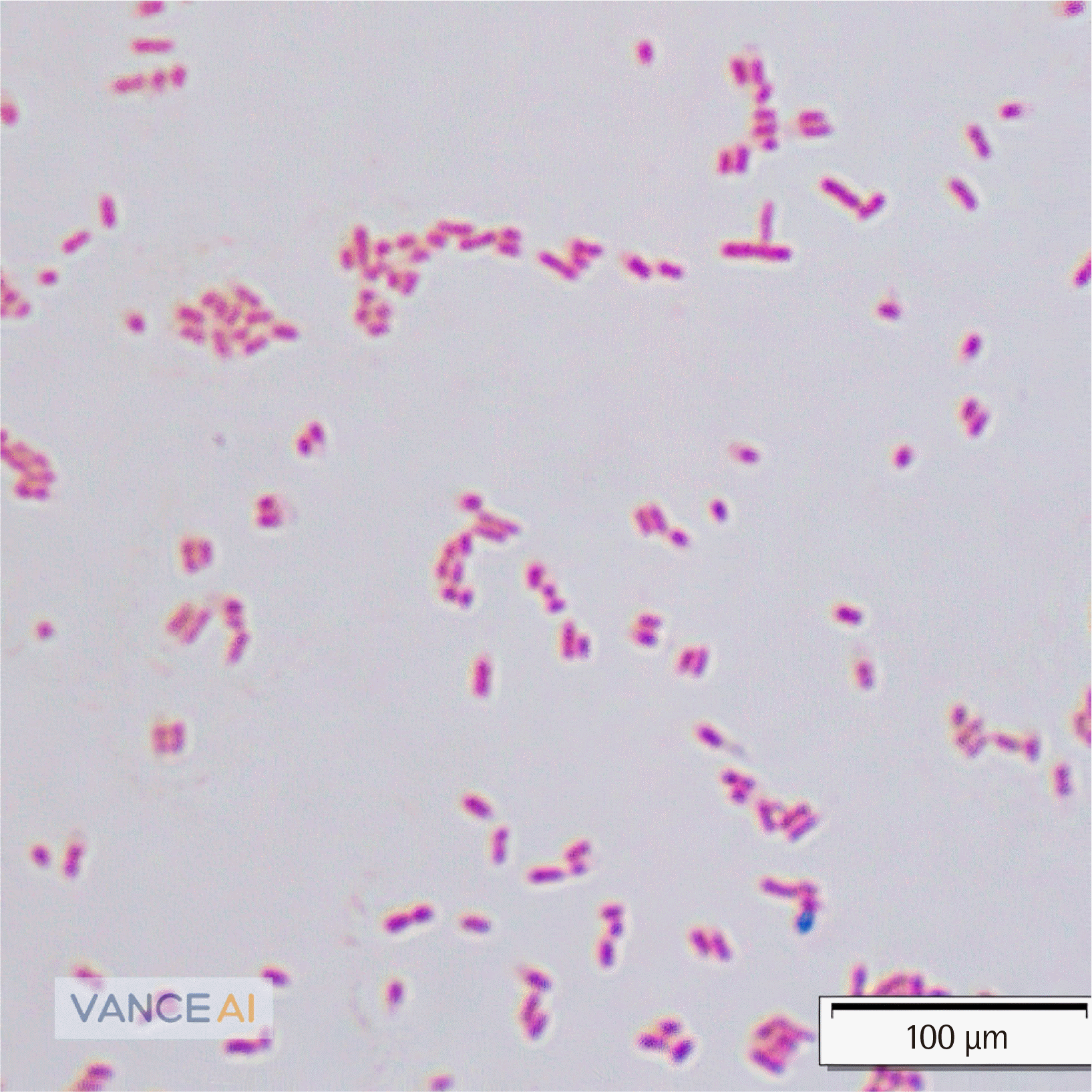
Table 1
Biochemical characteristics of Kosakonia cowanii
| Type strain 888-76 [5] | Current case | |
|---|---|---|
| Acid production | ||
| Adonitol | - | - |
| L-Arabitol | - | - |
| Cellobiose | + | + |
| Glucose | + | + |
| Maltose | + | + |
| Mannitol | + | + |
| Mannose | + | + |
| Sucrose | + | + |
| Sorbitol | + | + |
| Tagatose | - | - |
| Trehalose | + | + |
| Citrate | - | + |
| β-Galactosidase | + | + |
| β-Glucuronidase | + | - |
| β-Xylosidase | - | + |
| Indole | + | - |
| Lysine decarboxylase | - | - |
| Malonate | - | - |
| Ornithine decarboxylase | - | - |
| Urease | - | - |
Table 2
Comparison between the results of Kosakonia cowanii’s antibiotic susceptibility test in the current case and the results in previous reports
| Antibiotics | Berinson B [11] | Washio K [10] | Duployez C [3] | Current case | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||
| Etest with BMD | N/A | VITEK 2 | MicroScan | Etest | ||||||
|
|
|
|
|
|
||||||
| MIC | Susceptibility | MIC | Susceptibility | MIC | Susceptibility | MIC | Susceptibility | MIC | Susceptibility | |
| Ampicillin | ≥ 256 | R | > 16 | R | - | - | > 16 | R | - | - |
| Piperacillin | 8 | S | > 64 | R | 64 | R | 16 | SDD | - | - |
| Amoxicillin/clavulanic acid | - | - | - | - | ≤ 2 | S | > 16 | R | 1.5 | S |
| Ampicillin/sulbactam | ≤ 2 | S | < 8 | S | - | - | > 16 | R | - | - |
| Piperacillin/tazobactam | ≤ 0.5 | S | < 16 | S | - | - | ≤ 8 | S | 0.5 | S |
| Cefepime | - | - | < 2 | S | ≤ 1 | S | ≤ 1 | S | - | - |
| Cefotaxime | - | - | < 1 | S | ≤ 1 | S | ≤ 1 | S | < 1 | S |
| Ceftriaxone | - | - | < 1 | S | ≤ 1 | S | - | - | < 0.5 | S |
| Cefoxitin | 4 | S | - | - | - | - | > 16 | R | - | - |
| Cefuroxime | - | - | - | - | - | - | > 16 | R | - | - |
| Ceftazidime | - | - | < 4 | S | - | - | ≤ 1 | S | < 1 | S |
| Aztreonam | - | - | < 4 | S | - | - | ≤ 1 | S | - | - |
| Doripenem | - | - | - | - | - | - | ≤ 1 | S | - | - |
| Ertapenem | - | - | - | - | - | - | ≤ 0.5 | S | - | - |
| Imipenem | - | - | - | - | - | - | ≤ 1 | S | < 0.5 | S |
| Meropenem | ≤ 0.0032 | S | < 1 | S | - | - | ≤ 1 | S | < 0.25 | S |
| Colistin | ≤ 1 | I | - | - | - | - | ≤ 2 | I | - | - |
| Gentamicin | - | - | < 2 | S | - | - | ≤ 2 | S | < 0.5 | S |
| Tobramycin | - | - | - | - | - | - | ≤ 2 | S | - | - |
| Amikacin | - | - | < 4 | S | - | - | ≤ 8 | S | - | - |
| Tetracycline | - | - | - | - | - | - | ≤ 4 | S | - | - |
| Minocycline | - | - | < 2 | S | - | - | ≤ 4 | S | - | - |
| Ciprofloxacin | ≤ 0.04 | S | - | - | - | - | ≤ 0.5 | S or I | - | - |
| Levofloxacin | - | - | < 0.5 | S | - | - | ≤ 1 | S or I | < 0.5 | S |
| Trimethoprim/ | ||||||||||
| Sulfamethoxazole | - | - | < 2 | S | - | - | ≤ 2 | S | 0.125 | S |
| Chloramphenicol | - | - | - | - | - | - | ≤ 8 | S | - | - |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download