Abstract
T-lymphoblastic leukemia (T-ALL) is an aggressive hematologic malignancy associated with poor outcomes. The genetic background of T-ALL is widely heterogeneous and the TAL1 gene is overexpressed in approximately half of all cases. A submicroscopic interstitial deletion on chromosome 1p33 results in STIL-TAL1 fusion, causing inappropriate overexpression of TAL1, which promotes T cell leukemogenesis. T-ALL with STIL-TAL1 exhibits distinct characteristics, such as a mature cortical T cell immunophenotype, low incidence of NOTCH1 mutation, privileged association with PTEN inactivation, deletion of 6q14–q16, MYC translocation, high leukocyte count, poor response to treatment, and low event-free survival. However, the clinical relevance and prognostic value of this rearrangement remain unclear. Here, we report the first case of T-ALL with a 1p33 deletion resulting in STIL-TAL1 fusion in Korea, which was detected by reverse transcriptase-polymerase chain reaction and confirmed by chromosomal microarray analysis.
초록
T림프모구백혈병(T-ALL)은 공격적인 악성 혈액 종양으로 좋지 않은 예후를 보인다. T-ALL은 다양한 유전이상과 관련이 있으며, 약 절반 정도에서 TAL1 유전자의 과발현이 나타난다. 염색체 1p33의 미세 결실은 STIL-TAL1 재배열을 생성하고 이는 TAL1 유전자의 과발현을 유발하여, T 세포에서 백혈병으로 진행을 야기하게 된다. T-ALL에서 STIL-TAL1 재배열이 존재하는 경우, 성숙 T 세포의 면역표현형, 낮은 NOTCH1 유전자 돌연변이 발생률, PTEN 유전자 비활성화와의 연관성, 6q14-q16의 결실, MYC 유전자 전위, 진단 시의 높은 백혈구 수와 치료에 불량한 반응 등과 같은 독특한 특성을 보인다. 하지만, STIL-TAL1 재배열의 임상적 의의와 예후적 의미에 대해서는 아직 명확하게 알려져 있지 않다. 본 연구에서는 역전사효소-중합효소 연쇄반응(RT-PCR)과 염색체 마이크로어레이 분석(chromosomal microarray analysis)으로 확인한 1p33 결실로 인한 STIL-TAL1 재배열이 존재하는 T-ALL을 국내에서 첫 번째 증례로 보고하고자 한다.
T-lymphoblastic leukemia (T-ALL) is an aggressive hematologic malignancy that results from the malignant transformation of progenitors of T cells. T-ALL accounts for 10–15% of pediatric and 25% of adult ALL cases [1, 2] and is associated with a poor outcome [3]. The genetic background of T-ALL is widely heterogeneous as it is caused by the co-occurrence of multiple genetic abnormalities [1]. Therefore, molecular cytogenetic diagnosis is not currently adopted for T-ALL, and no integrated tests are available to clinically identify the broad spectrum of related genes and alternative molecular mechanisms [1].
TAL1 is a classic oncogene identified in chromosomal translocations in acute leukemia and is an important transcription factor that forms a key regulatory circuit [4]. A submicroscopic deletion on chromosome 1p33 results in STIL-TAL1 fusion (STIL is also known as SIL), causing inappropriate overexpression of TAL1, which promotes T cell leukemogenesis [3, 4]. STIL-TAL1 fusion has been suggested to be a likely founder or truncal event, with other common genetic lesions including CDKN2A loss, PTEN mutation or loss, and NOTCH1 mutation occurring as secondary subclonal events in STIL-TAL1 positive T-ALL [5]. However, the clinical relevance and prognostic value of this rearrangement remain unclear.
In this study, we identified STIL-TAL1 fusion in a patient with T-ALL using reverse transcriptase-polymerase chain reaction (RT-PCR) and confirmed a 1p33 deletion using chromosomal microarray analysis (CMA). To our knowledge, this is the first report of T-ALL with a 1p33 deletion resulting in STIL-TAL1 fusion in Korea.
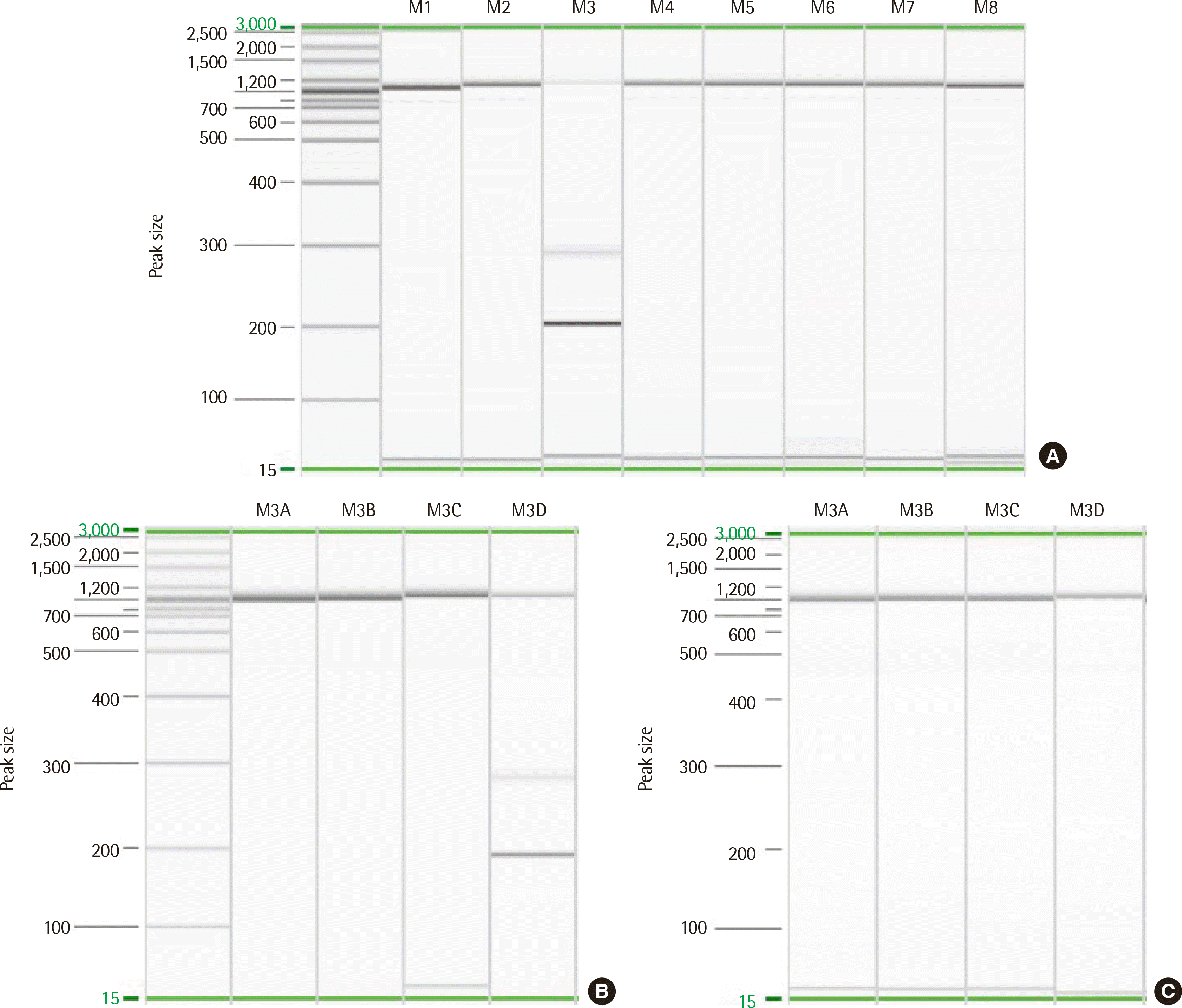
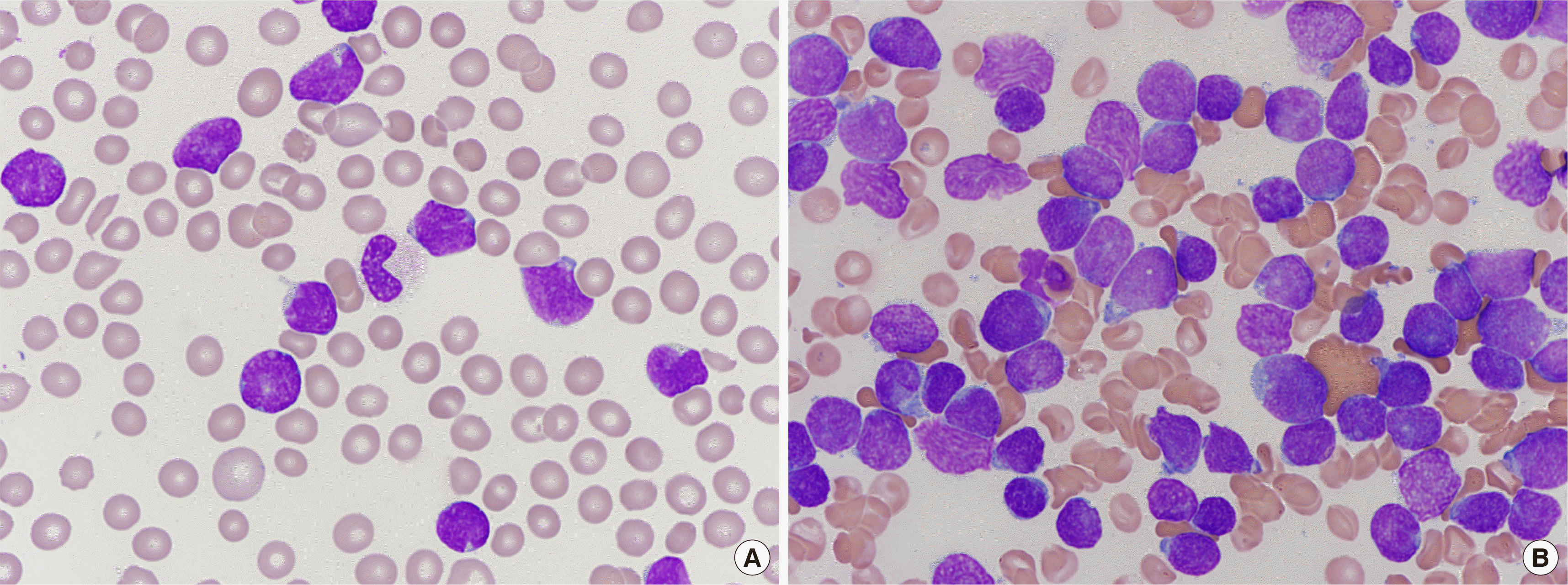
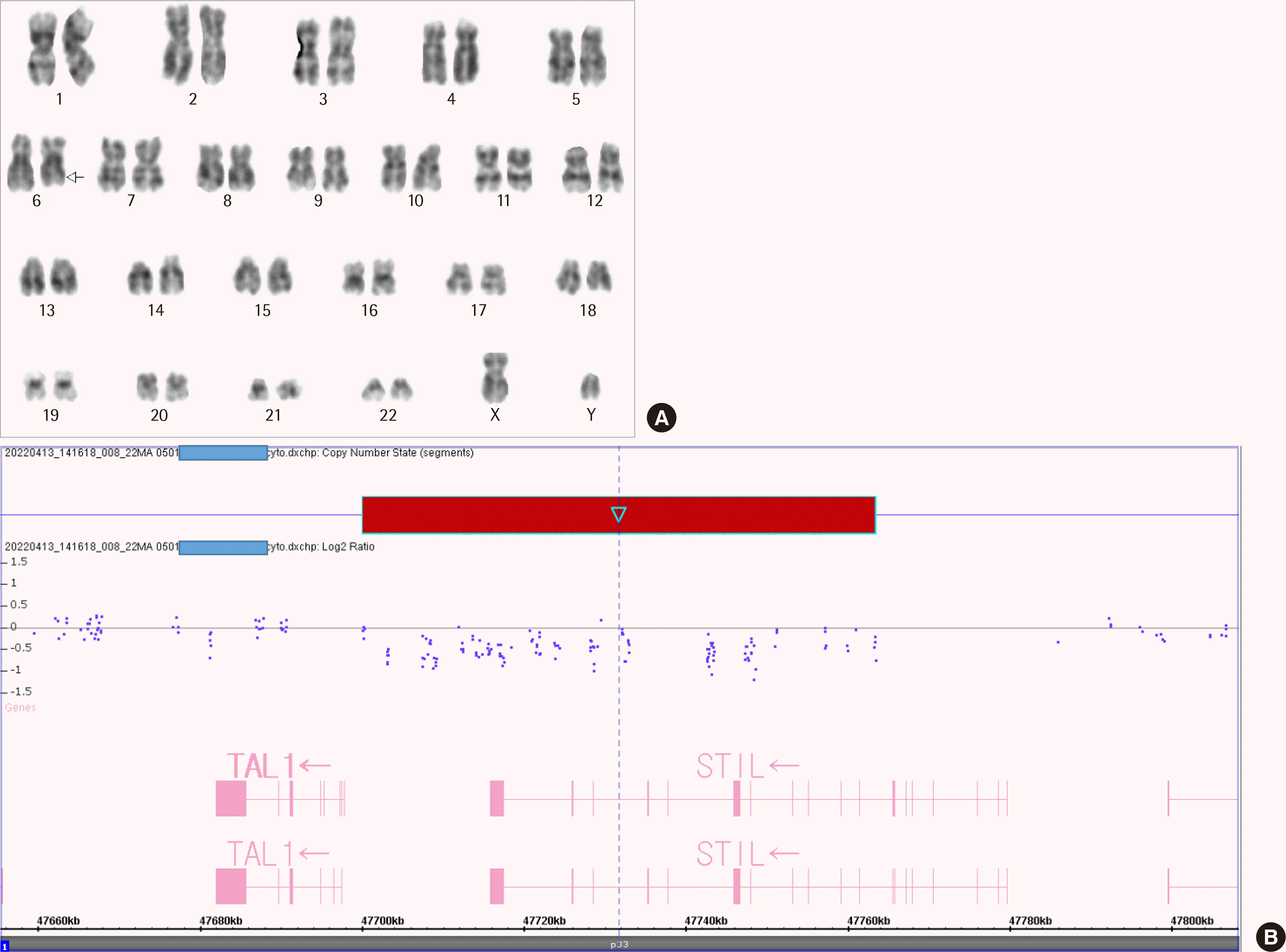
A 36-year-old man with chest discomfort and petechiae in both legs was referred to our emergency department. Laboratory results showed a leukocyte count of 197.6×109/L, hemoglobin level of 111 g/L, and platelet count of 16×109/L. Peripheral blood smears revealed 93% blasts (Fig. 1A). The bone marrow (BM) aspirate smear showed 93.5% blasts of small to medium size, with round or slightly cleaved nuclei (Fig. 1B). An immunophenotyping analysis revealed the blasts were positive for cytoplasmic CD3 (cCD3), CD3 (sCD3), CD5, CD7, CD38, and TdT. However, these cells did not express CD10, CD19, CD20, CD22, cCD22, cCD79a, CD13, CD33, CD14, CD64, CD117, or cytoplasmic MPO, suggesting T-ALL. Multiplex RT-PCR with HemaVision (DNA Diagnostic A/S, Risskov, Denmark) revealed a 191-bp STIL-TAL1 fusion transcript (Fig. 2A, B). The karyotype was 46,XY,del(6)(q21q23)[8]/46,XY[13] (Fig. 3A). Molecular analyses using Sanger sequencing revealed negative results for NPM1, CEBPA, FLT3-ITD/TKD, and Kit mutations. Next-generation sequencing (NGS) of DNA using an Illumina MiSeqDX platform and a custom-designed ALL panel revealed a NOTCH1 mutation, which is considered a variant of uncertain significance (VUS). Since the 1p33 deletion resulting in STIL-TAL1 fusion is a submicroscopic deletion, a microarray analysis was performed to confirm this. A CMA using the CytoScan DX Assay (Thermo Fisher Scientific Inc, Frederick, USA) based on the GRCh37/hg19 assembly showed a 63-kb heterozygous deletion at 1p33 (Fig. 3B) with additional copy number abnormalities, as follows: arr 1p33(47,700,108– 47,763,403)×1 [VOUS], arr 6q14.1q16.1(77,124,674–95,680,018)×1 [P], arr 7p14.1(38,293,949–38,395,208)×0 [VOUS], arr 7p34(142,016,358– 142,493,639)×1 [VOUS], arr 9p21.3(21,828,109–21,989,103)×0 [P], and arr 9p24.3p13.1(192,128–40,087,758)×2 hmz [VOUS]. Induction chemotherapy was administered to the patient. The follow-up study of BM one month later showed complete hematologic (0.6% blasts), cytogenetic (normal karyotype), and molecular remissions without STIL-TAL1 fusion transcript (Fig. 2C).
The TAL1 gene is overexpressed in approximately half of T-ALL cases as a result of chromosomal translocations, intrachromosomal rearrangements, or somatic mutations [4]. Translocation-induced TAL1 dysregulation is rarer than microdeletion-induced dysregulation. A submicroscopic interstitial deletion on chromosome 1p33 causes the 5′ untranslated region of STIL to fuse with the coding part of TAL1 [3, 4]. The deletion places TAL1 expression under the control of the STIL promoter, resulting in inappropriate overexpression of TAL1, which promotes T cell leukemogenesis [3, 4]. The STIL-TAL1 fusion gene has been reported in 15–25% of pediatric and young adult T-ALL cases, but much less frequently in older T-ALL patients [6]. A previous study in India found SIL-TAL1 fusion in 4/26 (15.4%) pediatric and none of the 7 adult T-ALL cases [7].
Chromosome deletions, as a mechanism for fusion gene formation, are mostly submicroscopic. Hence, they are not detected by cytogenetic analyses; however, they can be detected by array comparative genomic hybridization and/or high-throughput sequencing [6, 8]. In 1990, two independent research groups working on T-ALL used southern analysis to detect an approximately 90-kb interstitial deletion in 1p33 which caused STIL-TAL1 fusion, and this was the first description of a fusion gene resulting from an interstitial, submicroscopic deletion [6]. Yu et al. [8] performed a microarray analysis and found a submicroscopic deletion of approximately 80-kb in 1p33 (47,461,779–47,547,806 based on the hg18 assembly) leading to a STIL-TAL1 fusion in 9 of the 22 pediatric T-ALL patients. In the present case, the STIL-TAL1 fusion was first detected by multiplex RT-PCR. CMA confirmed a 63-kb deletion in 1p33 (47,700,108–47,763,403 based on the GRCh37/hg19 assembly).
Furthermore, T cell receptor rearrangement (deletion at 7p14.1 and 7q34) and biallelic deletion of CDKN2A (deletion at 9p21.3) were suspected based on the CMA results. NOTCH1 and CDKN2A play a surprising role, as they are altered in more than half of T-ALL cases [1, 9]. Activation of NOTCH1 signaling affects the specification and development of thymocytes, while CDKN2A haploinsufficiency/inactivation promotes cell cycle progression [1]. The present study revealed that PCR, NGS, and microarray analysis allow for a more comprehensive assessment of genetic abnormalities in T-ALL.
T-ALL with aberrations of TAL shares the immunophenotype indicative of arrest of mature cortical T cell development (sCD3 positive) and the same gene expression features as a low incidence of NOTCH1 mutation (~40% of cases), a privileged association with PTEN inactivation, deletion of 6q14–q16, and MYC translocation, all of which act as poor prognostic markers [1]. Previous studies have reported that STIL-TAL1 is associated with adolescence, higher leukocyte counts at diagnosis, poor response to treatment, and slightly lower event-free survival than T-ALL without STIL-TAL1 [3]. However, the clinical relevance and prognostic value of this rearrangement remain unclear, as different studies have reported favorable or unfavorable results in STIL-TAL1-positive patients [3, 6, 10]. The present case also exhibited distinct features similar to those mentioned previously, such as sCD3 expression, a NOTCH1 mutation, 6q14.1–6q16.1 deletion, and a very high leukocyte count at diagnosis. However, a follow-up study of BM one month later showed complete remission, indicating a good response to induction chemotherapy.
According to Zhao et al. [11], the STIL-TAL1 expression in 16 patients changed from negative before transplantation to positive at a median of 77 days (30–281 days) after transplantation, and 15 (93.8%) of them eventually experienced relapse. Their study demonstrated the reliability of STIL-TAL1 fusion transcripts as excellent minimal residual disease (MRD) markers. In the present study, expression of the STIL-TAL1 fusion transcript changed from positive at diagnosis to negative at complete remission a month later, supporting its role as a reliable marker for MRD.
In conclusion, we report the first case in Korea of T-ALL with a 1p33 deletion resulting in STIL-TAL1 fusion. More studies are required, including extensive cytogenetic and molecular analyses to identify novel therapeutic targets and to determine the pathophysiology, laboratory characteristics, clinical significance, and MRD monitoring of this gene rearrangement and other interstitial deletions/fusion genes.
REFERENCES
1. Bardelli V, Arniani S, Pierini V, Di Giacomo D, Pierini T, Gorello P, et al. 2021; T-cell acute lymphoblastic leukemia: biomarkers and their clinical usefulness. Genes. 12:1118. DOI: 10.3390/genes12081118. PMID: 34440292. PMCID: PMC8394887.
2. Van Vlierberghe P, Ferrando A. 2012; The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 122:3398–406. DOI: 10.1172/JCI61269. PMID: 23023710. PMCID: PMC3461904.
3. D'Angiò M, Valsecchi MG, Testi AM, Conter V, Nunes V, Parasole R, et al. 2015; Clinical features and outcome of SIL/TAL1-positive T-cell acute lymphoblastic leukemia in children and adolescents: a 10-year experience of the AIEOP group. Haematologica. 100:e10–3. DOI: 10.3324/haematol.2014.112151. PMID: 25304610. PMCID: PMC4281327.
4. Tan TK, Zhang C, Sanda T. 2019; Oncogenic transcriptional program driven by TAL1 in T-cell acute lymphoblastic leukemia. Int J Hematol. 109:5–17. DOI: 10.1007/s12185-018-2518-z. PMID: 30145780.

5. Furness CL, Mansur MB, Weston VJ, Ermini L, van Delft FW, Jenkinson S, et al. 2018; The subclonal complexity of STIL-TAL1+ T-cell acute lymphoblastic leukaemia. Leukemia. 32:1984–93. DOI: 10.1038/s41375-018-0046-8. PMID: 29556024. PMCID: PMC6127084.

6. Panagopoulos I, Heim S. 2021; Interstitial deletions generating fusion genes. Cancer Genomics Proteomics. 18:167–96. DOI: 10.21873/cgp.20251. PMID: 33893073. PMCID: PMC8126330.
7. Chopra A, Soni S, Verma D, Kumar D, Dwivedi R, Vishwanathan A, et al. 2015; Prevalence of common fusion transcripts in acute lymphoblastic leukemia: A report of 304 cases. Asia Pac J Clin Oncol. 11:293–8. DOI: 10.1111/ajco.12400. PMID: 26264145.

8. Yu L, Slovak ML, Mannoor K, Chen C, Hunger SP, Carroll AJ, et al. 2011; Microarray detection of multiple recurring submicroscopic chromosomal aberrations in pediatric T-cell acute lymphoblastic leukemia. Leukemia. 25:1042–6. DOI: 10.1038/leu.2011.33. PMID: 21383747.
9. Girardi T, Vicente C, Cools J, De Keersmaecker K. 2017; The genetics and molecular biology of T-ALL. Blood. 129:1113–23. DOI: 10.1182/blood-2016-10-706465. PMID: 28115373. PMCID: PMC5363819.
10. Wang D, Zhu G, Wang N, Zhou X, Yang Y, Zhou S, et al. 2013; SIL-TAL1 rearrangement is related with poor outcome: a study from a Chinese institution. PLoS One. 8:e73865. DOI: 10.1371/journal.pone.0073865. PMID: 24040098. PMCID: PMC3767609.
11. Zhao X, Hong Y, Qin Y, Xu Y, Chang Y, Wang Y, et al. 2017; The clinical significance of monitoring the expression of the SIL-TAL1 fusion gene in T-cell acute lymphoblastic leukemia after allogeneic hematopoietic stem cell transplantation. Int J Lab Hematol. 39:613–9. DOI: 10.1111/ijlh.12711. PMID: 28736882.
Fig. 2
Detection of the STIL-TAL1 fusion transcript resulting from 1p33 deletion by multiplex RT-PCR. The screening kit produced a single band in the M3 lane (A), and the split-out kit produced a single 191-base pair band in the M3D lane at diagnosis (B). The split-out kit revealed no band in the M3D lane in the follow-up study one month later (C).




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download