1. Tong Y, Xu S, Huang L, Chen C. Obesity and insulin resistance: pathophysiology and treatment. Drug Discov Today. 2022; 27:822–30.
2. Capurso C, Capurso A. From excess adiposity to insulin resistance: the role of free fatty acids. Vascul Pharmacol. 2012; 57:91–7.
3. Assimacopoulos-Jeannet F. Fat storage in pancreas and in insulin-sensitive tissues in pathogenesis of type 2 diabetes. Int J Obes Relat Metab Disord. 2004; 28 Suppl 4:S53–7.
4. Stenvers DJ, Scheer FA, Schrauwen P, la Fleur SE, Kalsbeek A. Circadian clocks and insulin resistance. Nat Rev Endocrinol. 2019; 15:75–89.
5. Lackey DE, Olefsky JM. Regulation of metabolism by the innate immune system. Nat Rev Endocrinol. 2016; 12:15–28.
6. Kim J, Lee J. Role of obesity-induced inflammation in the development of insulin resistance and type 2 diabetes: history of the research and remaining questions. Ann Pediatr Endocrinol Metab. 2021; 26:1–13.
7. Reinehr T. Inflammatory markers in children and adolescents with type 2 diabetes mellitus. Clin Chim Acta. 2019; 496:100–7.
8. Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, Spiegelman BM. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science. 1996; 271:665–8.
9. Ueki K, Kondo T, Kahn CR. Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol Cell Biol. 2004; 24:5434–46.
10. Ye J. Regulation of PPARgamma function by TNF-alpha. Biochem Biophys Res Commun. 2008; 374:405–8.
11. Bluher M. Adipose tissue inflammation: a cause or consequence of obesity-related insulin resistance? Clin Sci (Lond). 2016; 130:1603–14.
12. Shimobayashi M, Albert V, Woelnerhanssen B, Frei IC, Weissenberger D, Meyer-Gerspach AC, et al. Insulin resistance causes inflammation in adipose tissue. J Clin Invest. 2018; 128:1538–50.
13. Li Y, Yun K, Mu R. A review on the biology and properties of adipose tissue macrophages involved in adipose tissue physiological and pathophysiological processes. Lipids Health Dis. 2020; 19:164.
14. Michailidou Z, Gomez-Salazar M, Alexaki VI. Innate immune cells in the adipose tissue in health and metabolic disease. J Innate Immun. 2022; 14:4–30.
15. Russo L, Lumeng CN. Properties and functions of adipose tissue macrophages in obesity. Immunology. 2018; 155:407–17.
16. Giron-Ulloa A, Gonzalez-Dominguez E, Klimek RS, PatinoMartinez E, Vargas-Ayala G, Segovia-Gamboa NC, et al. Specific macrophage subsets accumulate in human subcutaneous and omental fat depots during obesity. Immunol Cell Biol. 2020; 98:868–82.
17. Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003; 112:1796–808.
18. Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007; 117:175–84.
19. Trim W, Turner JE, Thompson D. Parallels in immunometabolic adipose tissue dysfunction with ageing and obesity. Front Immunol. 2018; 9:169.
20. Villareal DT, Apovian CM, Kushner RF, Klein S; American Society for Nutrition; NAASO, The Obesity Society. Obesity in older adults: technical review and position statement of the American Society for Nutrition and NAASO, The Obesity Society. Obes Res. 2005; 13:1849–63.
21. Canning KL, Brown RE, Jamnik VK, Kuk JL. Relationship between obesity and obesity-related morbidities weakens with aging. J Gerontol A Biol Sci Med Sci. 2014; 69:87–92.
22. Tzanetakou IP, Katsilambros NL, Benetos A, Mikhailidis DP, Perrea DN. “Is obesity linked to aging?”: adipose tissue and the role of telomeres. Ageing Res Rev. 2012; 11:220–9.
23. Santos AL, Sinha S. Obesity and aging: molecular mechanisms and therapeutic approaches. Ageing Res Rev. 2021; 67:101268.
24. Greiss S, Gartner A. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol Cells. 2009; 28:407–15.
25. Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000; 273:793–8.
26. Gillum MP, Kotas ME, Erion DM, Kursawe R, Chatterjee P, Nead KT, et al. SirT1 regulates adipose tissue inflammation. Diabetes. 2011; 60:3235–45.
27. Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006; 444:868–74.
28. Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004; 429:771–6.
29. Schug TT, Xu Q, Gao H, Peres-da-Silva A, Draper DW, Fessler MB, et al. Myeloid deletion of SIRT1 induces inflammatory signaling in response to environmental stress. Mol Cell Biol. 2010; 30:4712–21.
30. Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARgamma. Mol Biol Cell. 2009; 20:801–8.
31. Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005; 280:13560–7.
32. Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell. 2013; 50:686–98.
33. Zaganjor E, Yoon H, Spinelli JB, Nunn ER, Laurent G, Keskinidis P, et al. SIRT4 is an early regulator of branched-chain amino acid catabolism that promotes adipogenesis. Cell Rep. 2021; 36:109345.
34. Shuai L, Zhang LN, Li BH, Tang CL, Wu LY, Li J, et al. SIRT5 regulates brown adipocyte differentiation and browning of subcutaneous white adipose tissue. Diabetes. 2019; 68:1449–61.
35. Kuang J, Zhang Y, Liu Q, Shen J, Pu S, Cheng S, et al. Fat-specific Sirt6 ablation sensitizes mice to high-fat diet-induced obesity and insulin resistance by inhibiting lipolysis. Diabetes. 2017; 66:1159–71.
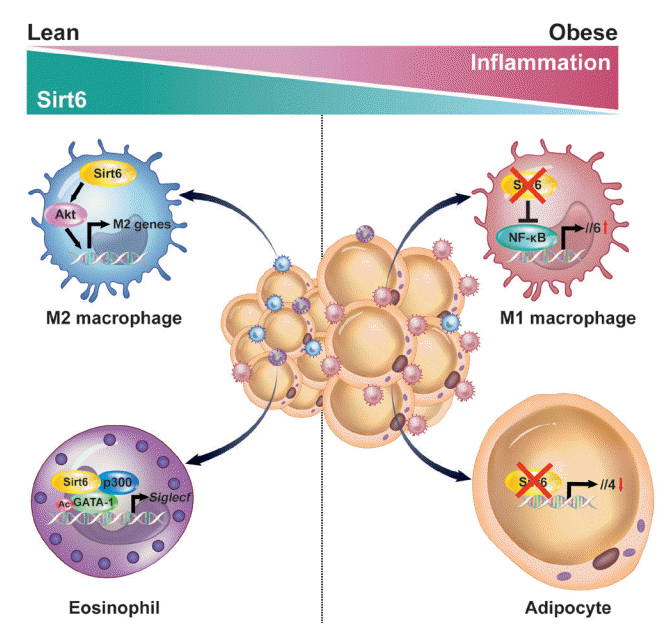
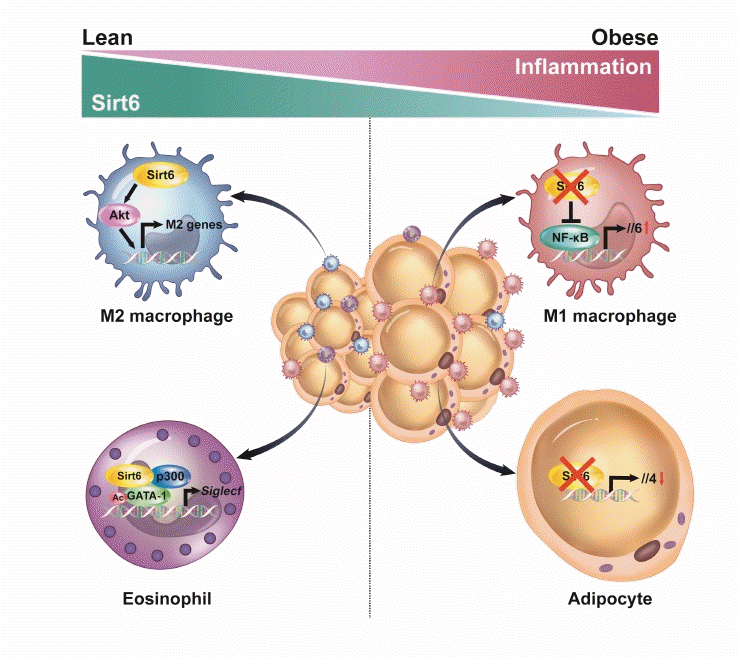
36. Song MY, Kim SH, Ryoo GH, Kim MK, Cha HN, Park SY, et al. Adipose sirtuin 6 drives macrophage polarization toward M2 through IL-4 production and maintains systemic insulin sensitivity in mice and humans. Exp Mol Med. 2019; 51:1–10.
37. Wu D, Bang IH, Park BH, Bae EJ. Loss of Sirt6 in adipocytes impairs the ability of adipose tissue to adapt to intermittent fasting. Exp Mol Med. 2021; 53:1298–306.
38. Xiong X, Zhang C, Zhang Y, Fan R, Qian X, Dong XC. Fabp4-Cre-mediated Sirt6 deletion impairs adipose tissue function and metabolic homeostasis in mice. J Endocrinol. 2017; 233:307–14.
39. Yao L, Cui X, Chen Q, Yang X, Fang F, Zhang J, et al. Cold-inducible SIRT6 regulates thermogenesis of brown and beige fat. Cell Rep. 2017; 20:641–54.
40. Fang J, Ianni A, Smolka C, Vakhrusheva O, Nolte H, Kruger M, et al. Sirt7 promotes adipogenesis in the mouse by inhibiting autocatalytic activation of Sirt1. Proc Natl Acad Sci U S A. 2017; 114:E8352–61.
41. Akter F, Tsuyama T, Yoshizawa T, Sobuz SU, Yamagata K. SIRT7 regulates lipogenesis in adipocytes through deacetylation of PPARγ2. J Diabetes Investig. 2021; 12:1765–74.
42. Qiao L, Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein alpha transcriptional complex. J Biol Chem. 2006; 281:39915–24.
43. Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X, et al. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007; 6:307–19.
44. Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008; 105:9793–8.
45. Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008; 8:333–41.
46. Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008; 8:347–58.
47. Chalkiadaki A, Guarente L. High-fat diet triggers inflammation-induced cleavage of SIRT1 in adipose tissue to promote metabolic dysfunction. Cell Metab. 2012; 16:180–8.
48. Hui X, Zhang M, Gu P, Li K, Gao Y, Wu D, et al. Adipocyte SIRT1 controls systemic insulin sensitivity by modulating macrophages in adipose tissue. EMBO Rep. 2017; 18:645–57.
49. Ka SO, Song MY, Bae EJ, Park BH. Myeloid SIRT1 regulates macrophage infiltration and insulin sensitivity in mice fed a high-fat diet. J Endocrinol. 2015; 224:109–18.
50. North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003; 11:437–44.
51. Li W, Zhang B, Tang J, Cao Q, Wu Y, Wu C, et al. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J Neurosci. 2007; 27:2606–16.
52. Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007; 6:505–14.
53. Krishnan J, Danzer C, Simka T, Ukropec J, Walter KM, Kumpf S, et al. Dietary obesity-associated Hif1α activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. 2012; 26:259–70.
54. Beirowski B, Gustin J, Armour SM, Yamamoto H, Viader A, North BJ, et al. Sir-two-homolog 2 (Sirt2) modulates peripheral myelination through polarity protein Par-3/atypical protein kinase C (aPKC) signaling. Proc Natl Acad Sci U S A. 2011; 108:E952–61.
55. Black JC, Mosley A, Kitada T, Washburn M, Carey M. The SIRT2 deacetylase regulates autoacetylation of p300. Mol Cell. 2008; 32:449–55.
56. Rothgiesser KM, Erener S, Waibel S, Luscher B, Hottiger MO. SIRT2 regulates NF-κB dependent gene expression through deacetylation of p65 Lys310. J Cell Sci. 2010; 123(Pt 24):4251–8.
57. Wang J, Koh HW, Zhou L, Bae UJ, Lee HS, Bang IH, et al. Sirtuin 2 aggravates postischemic liver injury by deacetylating mitogen-activated protein kinase phosphatase-1. Hepatology. 2017; 65:225–36.
58. Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010; 464:121–5.
59. Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011; 44:177–90.
60. Porter LC, Franczyk MP, Pietka T, Yamaguchi S, Lin JB, Sasaki Y, et al. NAD+-dependent deacetylase SIRT3 in adipocytes is dispensable for maintaining normal adipose tissue mitochondrial function and whole body metabolism. Am J Physiol Endocrinol Metab. 2018; 315:E520–30.
61. Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005; 280:21313–20.
62. Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009; 136:62–74.
63. Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, et al. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol. 2016; 23:434–40.
64. Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009; 8:2664–6.
65. Moschen AR, Wieser V, Gerner RR, Bichler A, Enrich B, Moser P, et al. Adipose tissue and liver expression of SIRT1, 3, and 6 increase after extensive weight loss in morbid obesity. J Hepatol. 2013; 59:1315–22.
66. Martinez-Jimenez V, Cortez-Espinosa N, Rodriguez-Varela E, Vega-Cardenas M, Briones-Espinoza M, Ruiz-Rodriguez VM, et al. Altered levels of sirtuin genes (SIRT1, SIRT2, SIRT3 and SIRT6) and their target genes in adipose tissue from individual with obesity. Diabetes Metab Syndr. 2019; 13:582–9.
67. Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006; 124:315–29.
68. Kanfi Y, Peshti V, Gil R, Naiman S, Nahum L, Levin E, et al. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010; 9:162–73.
69. Jang HY, Gu S, Lee SM, Park BH. Overexpression of sirtuin 6 suppresses allergic airway inflammation through deacetylation of GATA3. J Allergy Clin Immunol. 2016; 138:1452–5.
70. Lee Y, Ka SO, Cha HN, Chae YN, Kim MK, Park SY, et al. Myeloid sirtuin 6 deficiency causes insulin resistance in high-fat diet-fed mice by eliciting macrophage polarization toward an M1 phenotype. Diabetes. 2017; 66:2659–68.
71. Giblin W, Lombard DB. Sirtuin 6 builds a wall against inflammation, trumping diabetes. Diabetes. 2017; 66:2535–7.
72. Woo SJ, Noh HS, Lee NY, Cheon YH, Yi SM, Jeon HM, et al. Myeloid sirtuin 6 deficiency accelerates experimental rheumatoid arthritis by enhancing macrophage activation and infiltration into synovium. EBioMedicine. 2018; 38:228–37.
73. Koo JH, Jang HY, Lee Y, Moon YJ, Bae EJ, Yun SK, et al. Myeloid cell-specific sirtuin 6 deficiency delays wound healing in mice by modulating inflammation and macrophage phenotypes. Exp Mol Med. 2019; 51:1–10.
74. Iwasaki H, Mizuno S, Mayfield R, Shigematsu H, Arinobu Y, Seed B, et al. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. J Exp Med. 2005; 201:1891–7.
75. Willebrand R, Voehringer D. Regulation of eosinophil development and survival. Curr Opin Hematol. 2017; 24:9–15.
76. Bang IH, Park D, Lee Y, Cho H, Park BH, Bae EJ. Sirtuin 6 promotes eosinophil differentiation by activating GATA-1 transcription factor. Aging Cell. 2021; 20:e13418.
77. Rao RR, Long JZ, White JP, Svensson KJ, Lou J, Lokurkar I, et al. Meteorin-like is a hormone that regulates immune-adipose interactions to increase beige fat thermogenesis. Cell. 2014; 157:1279–91.
78. Qiu Y, Nguyen KD, Odegaard JI, Cui X, Tian X, Locksley RM, et al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell. 2014; 157:1292–308.
79. Huang Z, Zhong L, Lee JT, Zhang J, Wu D, Geng L, et al. The FGF21-CCL11 axis mediates beiging of white adipose tissues by coupling sympathetic nervous system to type 2 immunity. Cell Metab. 2017; 26:493–508.
80. Raj S, Dsouza LA, Singh SP, Kanwal A. Sirt6 deacetylase: a potential key regulator in the prevention of obesity, diabetes and neurodegenerative disease. Front Pharmacol. 2020; 11:598326.
81. Bae EJ. Sirtuin 6, a possible therapeutic target for type 2 diabetes. Arch Pharm Res. 2017; 40:1380–9.
82. Fiorentino F, Carafa V, Favale G, Altucci L, Mai A, Rotili D. The two-faced role of SIRT6 in cancer. Cancers (Basel). 2021; 13:1156.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download