INTRODUCTION
Deep learning (DL) is a type of machine learning-based analysis that employs a layered algorithmic architecture. Recent advances in DL have yielded marked success across various domains including, but not limited to, speech recognition, natural language processing, computer vision, and recommendation systems. In the healthcare domain, DL techniques are increasingly applied to medical image processing and analysis, natural language processing of large-scale medical text data, precision medicine, clinical decision support, and predictive analytics.
1 DL-based fast computation has dramatically reduced the time for genome analysis facilitating faster drug discovery and development.
23 The utilization of machine learning tools to predict disease has been increasingly described.
4
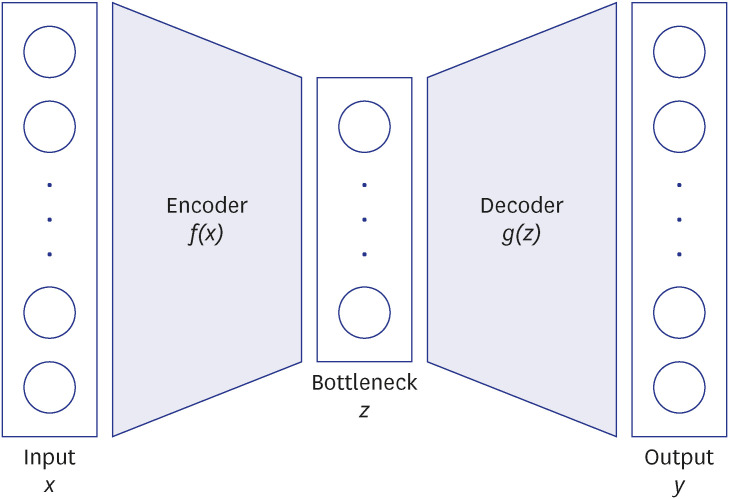
Autoencoder (AE) is one of the newer DL techniques that uses an artificial neural network to reconstruct its input data as an output while learning the latent encoding of unlabeled input data. AE’s capability in dimensionality reduction, feature extraction, and reconstruction of the data makes it a particularly effective DL technique to learn complex latent representation.
5 Leveraging this strength, one application of AE is DL-based recommender systems (RS) where RS can utilize latent features of recommended items (or latent user-item pair representation) which are learned by an AE. RS has gained popularity in commercial domains such as personalized movie recommendations in streaming services.
67
In healthcare, by analyzing patient-level data, RS can be applied to inferring diagnosis or recommending treatments.
89 RS can be extended to predictive modeling on a fixed problem such as predicting the relationship between the users and the given item. When built from a large patient dataset, DL-based RS can potentially allow healthcare providers to predict how likely a disease or diagnosis of interest would coexist or occur in the future in a given patient. Such information may prompt healthcare providers and patients to be more vigilant to screen for a disease of interest or to take preventative measures.
However, a challenge might stem from the fact that patient-level data is not uniformly and readily available, as they are highly subject to patient’s utilization of healthcare. For example, if a person rarely utilizes healthcare service, thereby not providing sufficient data input, the use of DL or other machine learning techniques, even if well trained and validated, prediction of disease or condition of interest would not be suitable. Further, even if data is available, complex logistics of accessing and pre-processing of raw healthcare data often make the DL application impractical. Given these challenges, our intention was to experiment the feasibility of prediction of the outcome of interest (disease of interest) using data that are more readily available.
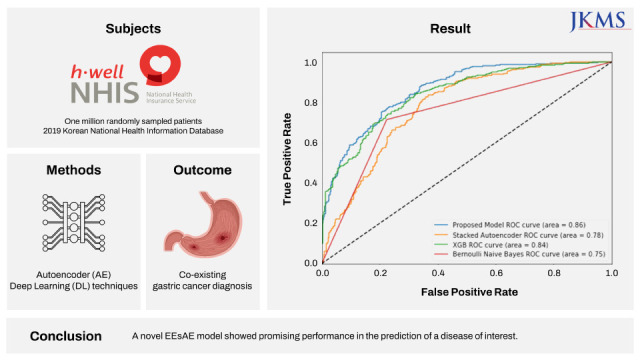
Specifically, we sought to demonstrate how the AE-based RS concept can be applied to predicting a disease of interest. In this study, we tested the hypothesis that gastric cancer (GC) can be predicted by AE-based RS solely based on other documented comorbid conditions available in the National Health Information Database (NHIS). The motivation for GC as a disease of interest was to investigate a disease that is common and relevant to the community from which data originates. GC is the second most prevalent cancer and is the third leading cause of cancer-related death in South Korea.
10 South Korea has the highest age-standardized rate per 100,000 of GC in the world. Approximately 244,000 new cases of GC were reported in South Korea in 2018.
11 We describe the construction and the performance of a novel AE-based DL model in the prediction of the GC diagnostic code using other diagnostic codes provided by the NHIS.
DISCUSSION
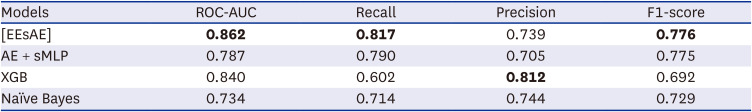
We demonstrated the construction and the performance of a novel supervised AE model in combination with MLP classifier in the prediction of disease of interest. The proposed model resulted in superior performances for the prediction of GC compared to other baseline models. Using documented ICD codes as the only input, the model yielded a recall of up to 0.82 in its identification of GC diagnosis. To the best of our knowledge, this is the first application of AE in the prediction of medical diagnosis based on diagnosis code inputs only.
Diagnostic codes are widely used in healthcare systems for the purpose of reimbursement, quality evaluation, public health reporting, and outcome research. Thus, a set of ICD codes typically represents a clinician’s culminating impression that results from various combinations of patient interviews, examination, and objective tests, such as laboratory or radiological studies. Despite the rich information that each ICD code may contain, the complex construct of how each ICD code is linked to other ICD codes requires DL process.
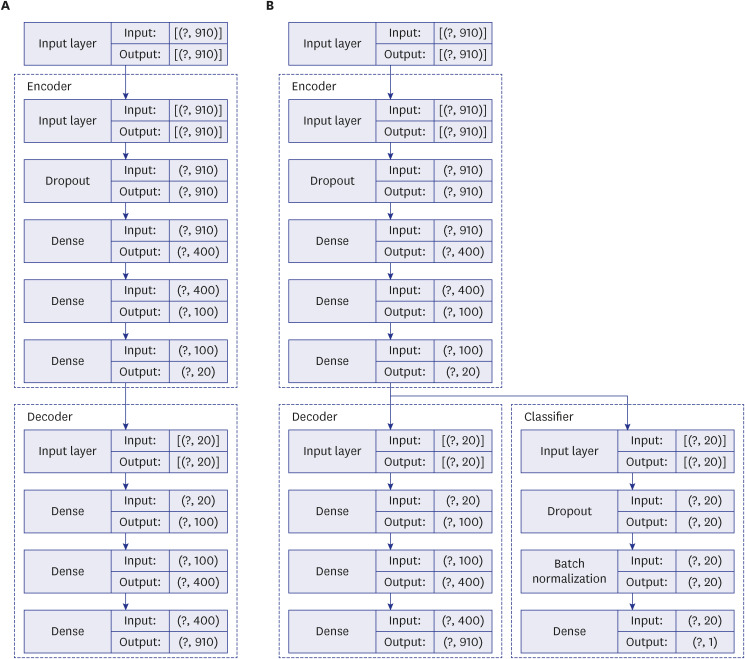
We posited that our proposed model would learn the complex construct and extract salient information sufficiently robust to predict the disease of interest. In doing so, we apply a RS concept of predicting relations of items and users to the problem of predicting the occurrence of disease (item) among patients (users). Herein, the medical history (neighboring ICD codes) represents the input for the RS and the disease of interest represents the fixed problem. The two AE models demonstrated herein are in line with a classic DL technique in which a model is fed with raw data and develops its own representations consisting of multiple layers of representations. Traditionally AE takes the form of unsupervised deep neural network model that learns the compression of the input data. We modified this by extending the AE in a supervised learning environment and combining with sMLP feature. Herein, the model takes advantage of each feature that learns to optimize the network. This enables better capturing of the complex latent representation of the raw data by lower-dimensional representations. We further refined the model by updating the classified and the AE simultaneously in an end-to-end fashion aiming to minimize custom loss function from both the AE and MLP.
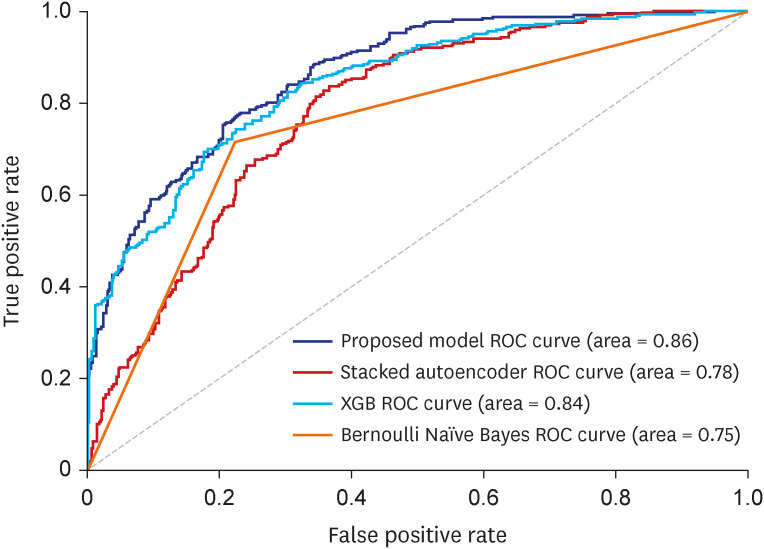
The superior performance of our proposed model in recall over baseline models points to the effectiveness of lower-dimensional representations. The AE mapped similar ICD codes into the low dimensional latent space where they are close to each other, and let the classifier discover patterns for predicting the target disease of interest, GC in this study. We also found when the AE and MLP are updated simultaneously as in our proposed EEsAE, the latent representation was even better learned and led to superior performances. The demonstration of F1 score of 0.78, recall of 0.82, and AUC of 0.86 epitomizes the potential of its application in similar predictions. Recall function (sensitivity) is a more relevant metric in prediction of a disease of interest, particularly where screening is important. This implies that about 4 out of 5 people with GC would be detected by the model using coexisting ICD codes alone. One can speculate that such a predictive model can help clinicians raise a high index of suspicion for a certain disease of interest, which is in this case, GC. For example, the model can prompt a clinician to perform more proactive screening of GC. Alternatively, the model may lead to more proactive prevention efforts.
Currently, screening for GC is controversial, and recommendations for screening is dependent on the incidence of GC. In a country where the incidence of GC is excessively high, such as Korea, universal population-based screening is being implemented (e.g., upper endoscopy every 2 years) for individuals aged 40 to 75 years. However, in many other countries where the incidence of GC is low, selective screening of high risk subgroups is advocated. Such high risk groups include those with gastric adenomas, pernicious anemia or familiar adenomatous polyposis, some of which require endoscopy. In that sense, the findings of our study provide a glimpse into how DL, specifically an AE model, might help identify potentially high risk group of patients of GC, who may benefit from more proactive screening with endoscopy especially where the incidence of GC is low.
Prior works have indeed used AE for medical diagnosis.
1819 However, the majority of these prior studies utilize AE analysis of medical imaging rather than ICD code. Our model, particularly the use of the EEsAE model, and its application to diagnosis just using ICD code is a first to our knowledge. In the testing phase in the evaluation of our model, all the occurrences and non-occurrences of GC code have been hidden. The disease codes have been aggregated by each patient ID without the information of temporal sequence in occurrences. Therefore, such an inference is still considered prediction. The prediction model is still meaningful in two ways. First, our goal was to better understand the similarity across different diagnostic codes, not the causal relationship in reference to the GC diagnostic code. Given the major clinical implication of the diagnosis like GC, and the challenge in early diagnosis, an AE model that “recommends” GC in this setting can be valuable for clinicians. Second, despite not being able to consider the temporal sequence due to the nature of the data, it actually reflects real world clinical practice where diagnosis is often delayed or even missed. Indeed, the timing of the diagnosis (i.e., first appearance of diagnostic codes) does not equate to the timing of the true onset of the condition.
Despite the potential implication, the findings of the study should be interpreted in the context of how the data was pre-processed. Since a model construction involved only people with at least 6 ICD codes with each code found in at least more than 50 people in the dataset were included, model performance is expected to be poor in people with the scarce number of ICD codes or with relatively rare ICD codes. Moreover, we validated our data in the subcohort from the same year, but have not tested the performance in data from a different year due to the limited availability of the data. Future studies should examine whether a similar level of prediction can be achieved when the model is applied to dataset from another year and for another disease of interest from the data of the same year, as well as different years.
Another limitation of the dataset used in this study is inherent to the limitation of the administrative data. The validity of ICD code varies across different conditions and healthcare systems and depends on how data is collected.
20 ICD based diagnosis information is not comprehensive. Moreover, an ICD code does not necessarily reveal the onset of the condition. Code assignment is a demanding process with many potential sources of errors.
21 Moreover, ICD code assignment practice is variable among clinicians.
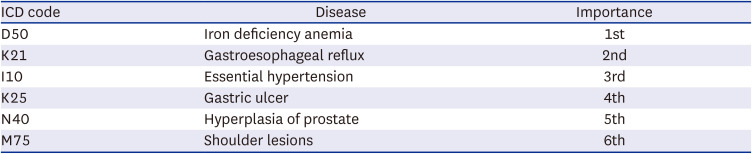
It is equally important to understand the inherent challenge in the interpretation of the results beyond the prediction. While some of the top influencing ICD codes identified in the ablation study appear to have associations with GC, the exact relationship is difficult to interpret. For example, the ablation study revealed IDA as the top influencer in the model meaning that the largest performance reduction was noted when this particular ICD code was excluded. While it seems plausible that there might be a link between the IDA and GC possibly via loss of blood in gastrointestinal track in the setting of gastric pathology, such a link has rarely been documented. One recent study revealed the high prevalence of IDA up to 40% at the time of GC diagnosis.
22 Among those who received chemotherapy for GC about 20% developed IDA along their treatment course.
23 Prior studies in Korea have simply reported the prevalence of IDA in patients with GC before and after gastrectomy.
24 Despite the potential link, specifically how this particular ICD code stood out as the top influencing variable outperforming other variables is unclear. GERD is most closely related to esophageal adenocarcinoma and to a lesser extent to GC arising from the cardia of the stomach.
25 Since only the main category of GC, C16, was used without consideration of subcategory information, such as area of the cancer, it is impossible to further speculate how GERD is linked to GC. Unlike other predictors, an association between gastric ulcer and GC can be more readily inferred through common risk factors (i.e., mainly
Helicobacter pylori infection).
26 Essential hypertension, benign prostate hyperplasia, and shoulder lesions are rather unanticipated diagnoses with little known association with GC. It is very possible that these entities are somehow linked through their associations with common risk factors, confounders, or the consequences of GC treatment. It is important to note that these factors would be more pertinent to the population where the model was trained. Nonetheless, elucidation of influencing factors can potentially enable researchers to explore associations of unanticipated factors with the disease of interest.
This study represents a benchmarking framework for future studies in which additional patient-level characteristics can be added to further enhance the model performance. For example, while this study only included ICD codes in the model, various data including demographics, medical history beyond the diagnosis, and test results can be included to construct even higher performing models.
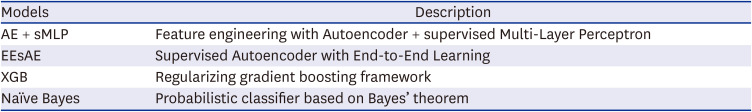
In conclusion, we describe a novel supervised AE in its application for the prediction of disease of interest. We showed that neighboring ICD code information alone predicted the co-existence of GC with high accuracy. Specifically, the proposed EEsAE model and AE-sMLP outperformed XGB and Naïve Bayes models. While the utility of the AE methods in the prediction of disease of interest (vs. identification of coexisting disease of interest) needs to be evaluated in a prospective study design, this high performance of the proposed model encourages us to further explore its utility and performance in other healthcare domains including future disease prediction.




 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download