Abstract
Background
Microtubule actin crosslinking factor 1 (MACF1) is a spectraplakin cytoskeletal crosslinking protein whose function and role in cancer biology has lacked investigation. Recent studies have identified MACF1 as a novel target in glioblastomas expressed in tissue from tumor patient explants but not normal brain tissue and when silenced has an antitumorigenic impact on these tumors. Radiation as a single agent therapy to treat glioblastomas has been used for decades and has done little to improve survival of individuals diagnosed with this disease. However, contemporary clinical radiotherapy protocols have provided evidence that combinatorial radiotherapy approaches confer a therapeutic benefit in glioblastoma patients. In this study MACF1 was investigated as a radiosensitization target in glioblastomas.
Methods
To provide context of MACF1 in glioblastomas, The Cancer Genome Atlas expression analyses were performed in conjunction with genes associated with glioblastoma evolution, while a genetic inhibitory approach, cell migratory assays, and immunofluorescence procedures were used to evaluate responses to MACF1 suppression with radiation. Additionally, expression analyses were conducted to assess co-expression of mTOR signaling pathway regulators and MACF1 in glioblastoma patient samples.
Results
Our amalgamation approach demonstrated that negative regulation of MACF1, which was positively correlated with epidermal growth factor receptor and p70s6k expression, enhanced the sensitivity of glioblastoma cells to radiation as a consequence of reducing glioblastoma cell viability and migration. Mechanistically, the antitumorigenic effects on glioblastoma cell behaviors after radiation and impairing MACF1 function were associated with decreased expression of ribosomal protein S6, a downstream effector of p70s6k.
Conclusion
MACF1 represents a diagnostic marker with target specificity in glioblastomas that can enhance the efficacy of radiation while minimizing normal tissue toxicity. This approach could potentially expand combinatorial radiation strategies for glioblastoma treatments via impairment of translational regulatory processes that contribute to poor patient survival.
Microtubule actin crosslinking factor 1 (MACF1) is a member of the plakin protein family discovered a little over two decades ago as a cytoskeletal cross-linking protein between actin microfilaments and microtubules that has been shown to contribute to cell proliferation and migration across diverse cellular systems [1]. More specifically it was demonstrated that MACF1 was prominently involved in neuronal cell migration [2], while subsequent investigation by Hu et al. [3], provided evidence that MACF1 when knocked down compromised osteoblasts proliferation. Despite established roles of MACF1 in cell behaviors that contribute to the propagation and maintenance of cancers, few studies have evaluated it as a therapeutic target in this disease. To this end we recently provided evidence that MACF1 was expressed at high levels in glioblastomas but not in lower grade brain tumors and normal brain tissue [4]. Additionally, down-regulation of MACF1 which has been described to have genetic mutations in renal cell carcinomas and endometrial cancers [56], had an anti-tumorigenic effect on glioblastoma cell viability and migration [4]. The inhibition of protumorigenic glioblastoma cell behaviors as a consequence of silencing MACF1 function were associated with down-regulation of Wnt signaling mediators, Axin1 and beta-catenin [4]. These data were consistent with findings in a seminal study by Chen et al. [7], which also described the interaction of MACF1 with Wnt signaling mediators, and are significant because Wnt signaling is a known mechanistic pathway that contributes to the progression of several cancers including glioblastomas [8]. From a therapeutic standpoint we also demonstrated that temozolomide, the gold standard chemotherapeutic drug used to treat glioblastomas induced MACF1 expression in these tumors and had a synergistic effect when combined with MACF1 inhibition [4].
Expanding upon our findings, we evaluated here the effects of inhibiting MACF1 as a means to enhance and sensitize the response of glioblastomas to radiation therapy in in vitro glioblastoma model systems. Radiation is typically used clinically to treat patients diagnosed with glioblastomas following surgical resection. An unfortunate caveat has been the limited efficacy of radiation as a single treatment option that enhances overall survival of patients diagnosed with this disease [910]. However, pioneering work by Stupp et al. [11], established a clinical precedent for the utility of combinatorial radiation therapy approaches for the treatment of glioblastomas, when they demonstrated an enhanced therapeutic benefit to patients that received radiation plus the chemotherapeutic agent temozolomide, as compared to patients that received radiation treatment alone. To date however, the primary targets of combinatorial radiotherapy approaches in glioblastomas have been limited to DNA repair proteins and protein kinase signaling cascades [1213141516]. Inhibitory targeting of MACF1 as a radiosensitizer, represents a novel experimental strategy that broadens combinatorial radiotherapy approaches in genetically heterogeneous glioblastomas that is essential to improving and managing disease progression. Additionally, we examined and identified ribosomal protein S6, a pro-tumorigenic downstream signaling mediator in the mTOR pathway [1718] and whose expression has been associated with poor survival of glioblastoma patients [19] as a mechanistic contributor of the combinatorial impact of MACF1 inhibition and radiation treatment.
U251 human glioblastoma cells were purchased from Sigma-Aldrich (St. Louis, MO, USA; 09063001) and A172 human glioblastoma cells from the American Type Culture Collection (Manassas, VA, USA; ATCC-CRL 1620). All cell lines were maintained in Dulbecco's Modified Eagles Medium-DMEM (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), 2 mM L-glutamine (Invitrogen), 100 nM MEM non-essential amino acids (Invitrogen), and penicillin-streptomycin (Invitrogen) at 37℃ and 5% CO2. GIPZ lentiviral shRNAs were purchased from Dharmacon (Chicago, IL, USA) and a Mark 1 Cesium-137 source was used to treat cells with a single 5 Gy dose of radiation in the Department of Radiation Oncology at Vanderbilt University Medical Center [20].
shRNA lentiviral transduction was performed with one of three lentiviral shRNAs targeting MACF1 (1-V2LHS_28596; 2-V3LHS_306210; 3-V3LHS_306213-3) in 1×105 U251 and A172 cells in serum free media with a multiplicity of infection (MOI) of 0.9 overnight at 37℃ and 5% CO2; cells were transduced with non-silencing shRNA as a control. Next complete growth media was added to U251 cells containing lentiviral shRNAs and allowed to incubate for 3–4 days. Following initial transduction of A172 cells, serum free media containing lentiviral shRNAs was replaced with normal growth media for 24 hours. Subsequently, growth media was removed and replaced with normal growth media containing 187 ng/mL of puromycin and incubated for 72 hours. Cells were then trypsinized, replated, and incubated in fresh growth media with puromycin for an additional 24 hours prior to treatment with radiation. MACF1 expression was subsequently examined in cells treated with control shRNAs and cells transduced with shRNAs targeting MACF1 using immunofluorescence procedures. Experiments were each performed at least three times. Means were determined between three separate experiments and statistical analysis was performed using Student's t-test to evaluate significance between experimental conditions using Graphpad Prism 8 (GraphPad Software, Inc., San Diego, CA, USA).
U251 and A172 cells were either treated with shRNAs alone, irradiated with 1×5 Gy, or treated with shRNAs (Dharmacon, Chicago, IL, USA) prior to irradiation as described above. Subsequently, 7×103 cells were plated and allowed to incubate for 1, 2, 3, or 5 days in DMEM (Invitrogen) containing 10% fetal bovine serum (Invitrogen). A172 cells were incubated in media containing puromycin. At the end of each time point tissue culture media was removed, the cell monolayer fixed with 100% methanol for 5 minutes and stained with 0.5% crystal violet in 25% methanol for 10 minutes. Cells were then washed three times 5 minutes each with distilled water to remove excess dye and allowed to dry overnight at room temperature. Cells were then manually counted or the incorporated dye was solubilized in 0.1 M sodium citrate (Sigma-Aldrich) in 50% ethanol. Next, 100 µL of experimental samples were transferred to 96 well plates and optical densities read at 595 nm using a X-mark microplate absorbance spectrophotometer (BioRad, Hercules, CA, USA). Experiments were each performed at least three times with duplicate samples and means determined by averaging duplicate samples within each independent experiment. ANOVA followed by Tukey's posthoc analysis were used to evaluate significance between means of experimental conditions using Graphpad Prism.
Motility assays were conducted according to manufacturer's instructions (CellBiolabs Inc., San Diego, CA, USA). Media containing 10% fetal bovine serum was added to the lower chamber of the migration well, while 2×104 irradiated and shRNA treated irradiated cells were prepared in 300 µL of serum free media, added to the Boyden chamber insert, and allowed to incubate for 24 hours at 37℃ and 5% CO2. Subsequently, non-migratory cells were removed from plate inserts (per manufacturer's instructions) and migratory cells stained with proprietary staining solution. Images were captured using an Olympus IX53 (Olympus, Center Valley, PA, USA) microscope and optical densities read at 560 nm in proprietary solubilization solution using a X-mark microplate absorbance spectrophotometer (BioRad). Experiments were each performed at least three times with duplicate samples and means determined by averaging duplicate samples within each independent experiment. ANOVA followed by Tukey's post-hoc analysis were used to evaluate significance between means of experimental conditions using Graphpad Prism.
For immunofluorescence labeling 1.5×104 cells were plated that were naïve controls, treated with shRNAs alone, radiation alone, or the combination of shRNAs and radiation. Cells were rinsed in PBS, and fixed in 4% paraformaldehyde for 5 minutes at room temperature. Next cells were rinsed with PBS post-fixation, permeabilized in 0.075% Triton X-100/PBS for 5 minutes, rinsed again with PBS, and blocked with 3.0% bovine serum albumin, 1.5% horse serum (Vector Labs, Burlingame, CA, USA) in PBS for 1 hour at room temperature. Cells were then incubated overnight at 4℃ with antibodies that detect MACF1 (377534; Santa Cruz Biotechnology, Dallas, TX, USA) and p-S6 ribosomal protein (4858; Cell Signaling Technology, Danvers, MA, USA). Samples were then rinsed three times with PBS, incubated with an Alexa 488 goat-anti-mouse conjugated secondary antibody (4412; Cell Signaling Technology) for 1 hour in the dark, rinsed again and examined with an Olympus IX53 microscope. Quantification of fluorescence was determined by measuring the fluorescence intensity from a total of 50 individual cells from three separate independent experiments and averaged. Statistical analysis was performed using ANOVA followed by Tukey's post-hoc analysis to evaluate significance between means of experimental conditions using Graphpad Prism. Antibody validation was performed via western blotting that assessed molecular weight and specificity of target proteins. Experiments were each performed at least three times.
In an effort to expand findings from our previous investigation that showed MACF1 was expressed in glioblastoma tissue but not in normal brain tissue [4], The Cancer Genome Atlas (TCGA-cbioportal) was used to gain further insight regarding the association of MACF1 expression and glioblastoma patient survival [21]. A comparative survival curve analysis was performed on patient cohorts stratified by MACF1 expression and showed a relative decrease of patient survival in glioblastomas with high MACF1 expression as compared to low MACF1 expressing tumors between 5–15 months (Fig. 1). Additionally, correlative expression analyses between MACF1 and aberrantly expressed genes known to contribute to the etiology and progression of these tumors revealed a statistically significant positive correlation between MACF1 and epidermal growth factor receptor (EGFR) expression, while a negative correlation was observed between MACF1, PTEN, CDKN2A, and TP53 expression (Fig. 1).
At present there are no pharmacological inhibitors to impair MACF1 cross-linking and signaling functions. To this end we have previously shown that genetic inhibition of MACF1 reduced glioblastoma cell proliferation in established and patient derived xenograft glioblastoma cell lines, which provided the technical feasibility and foundation for the present study [4]. Therefore, MACF1 expression and function here were down-regulated in glioblastoma cells using a lentiviral transduction shRNA genetic inhibitory approach that induced a considerable reduction in the cytoplasmic expression of this cytoskeletal protein (Fig. 2). Reduced MACF1 protein expression to shRNAs targeting MACF1 was accompanied by decreased glioblastoma cell viability. Specifically, a 28% and 48% decrease in U251 cell viability was observed in response to shRNAs targeting this cytoskeletal cross-linker as compared to non-targeting shRNA controls, while shRNAs targeting MACF1 in A172 cells had a 50% and 17% reduction in cell viability when compared to non-targeting controls (Fig. 3).
As a single treatment modality radiation has not been clinically beneficial in improving the overall survival of glioblastoma patients [23]. This is a consequence of intrinsic and acquired radiation resistance that can be attributed to genomic instability and radiation induced expression of DNA damage response proteins [222324] that contribute to the invasive nature of these tumors and their inevitable recurrence [2526272829]. Consistent with this notion cbioportal data showed that MACF1 positively correlated with mRNA expression of several DNA damage repair proteins in clinical glioblastoma patients (Table 1). Experimentally, the combinatorial efficacy of radiation and down-regulation of MACF1 protein expression had a demonstrative effect on glioblastomas, with as much as an 82% (p<0.01) and 32% (p<0.05) statistically significant decrease in U251 and A172 cell viability, respectively, when compared to non-targeting shRNA control cells treated with radiation (Fig. 3). Additionally, the combinatorial effects of radiation and MACF1 inhibition on the migratory capacity of glioblastoma cells were determined using Boyden chamber cell migration assays. These data revealed that U251 and A172 cell migration was abolished in cells treated with shMACF1-2 and radiation, albeit A172 glioblastoma cell migration was more susceptible to combination treatments (p<0.05; p<0.01) as compared to controls (Fig. 4). It should also be noted that the anti-migratory response of glioblastoma cells to the combination of radiation and MACF1 inhibition is not likely a consequence of cell death, as a decrease in the number of viable cells was not observed at time points relative to the number of viable cells at the day zero time point (data not shown).
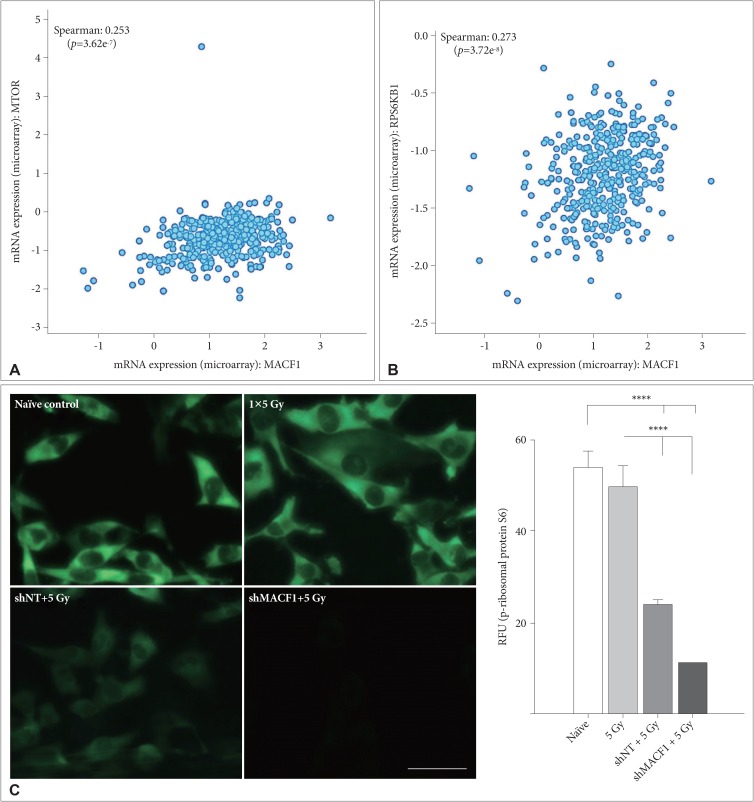
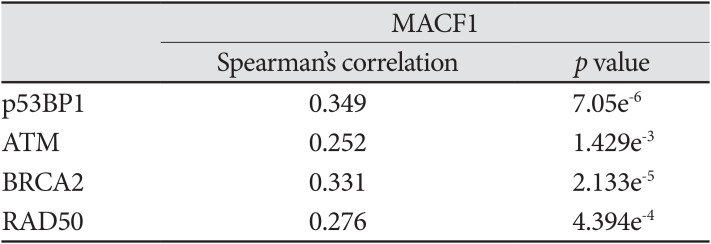
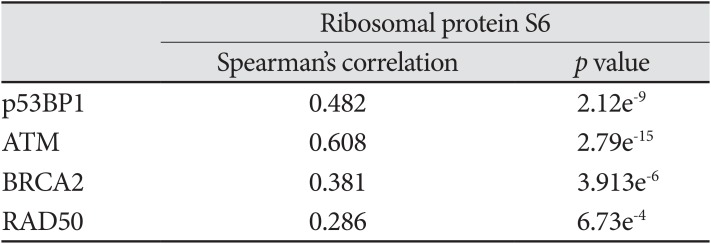
Mechanistic evaluation of the radiosensitization effects of impairing MACF1 in glioblastoma cells was assessed by evaluating changes in ribosomal protein S6 of the mTOR signaling pathway, a contributor in tumor cell proliferation and metastatic invasion. Like MACF1, ribosomal protein S6 had a positive correlative expression with several DNA damage repair genes in clinical glioblastoma patients (Table 2). Effector molecules of ribosomal protein S6, mTOR and its immediate downstream target, RPS6KB1 (p70s6k), whose expression have been associated with shorter overall survival of glioblastoma patients [30] were positively correlated with MACF1 in a cohort of patients (Fig. 5). Additionally, the activated form of ribosomal protein S6 that is regulated by p70s6k was also assessed as a mechanistic contributor of the radiosensitization effects of MACF1 inhibition. Although mRNA co-expression analysis did not reveal a positive correlation between MACF1 and ribosomal protein S6, an association of MACF1 and activated ribosomal protein S6 as a mechanistic signaling mediator in glioblastomas was observed here. To this end an 81% and 80% decrease of p-ribosomal protein S6 expression (p<0.0001) was seen in U251 cells treated with radiation and shMACF1 as compared to naïve untreated controls and cells treated with radiation alone (Fig. 5).
Diagnostic biomarkers as targets and predictors of radiotherapy response for the clinical treatment of glioblastomas have been somewhat ambiguous and limited to the presence of genetic alterations of the EGFR and methylation status of O-6-methylguanine DNA methyltransferase (MGMT) [313233343536]. However, genetic and cellular glioblastoma heterogeneity warrant the identification of expanded targets that can improve overall survival outcomes. We previously demonstrated that the spectraplakin protein, MACF1, is a potential novel diagnostic therapeutic target in glioblastomas and when negatively regulated induces an antitumorigenic response to the propagation and progression of these tumors [4]. In furtherance of this, we showed here that MACF1 enhanced and sensitized glioblastoma cells to radiation via a synergistic combinatorial effect when genetically silenced. These findings parallel a recent preclinical investigation of the microtubule poison, eribulin that concomitantly sensitized glioblastomas to radiation in an orthotopic xenograft model and established along with data here that cytoskeletal proteins continue to be a viable radiosensitization target in these solid tumors [37].
An additional consideration as part of the therapeutic management of glioblastomas are their high recurrence rates as a consequence of invading local normal tissue. To this end it was also demonstrated that the combination of MACF1 silencing and radiation reduced the migratory ability of these tumor cells, further enhancing the salubrious impact of this radiosensitization approach. The anti-tumorigenic effects of radiosensitizing glioblastoma cells by inhibiting MACF1 was also accompanied by reduced expression of ribosomal protein S6, a downstream effector of the mTOR signaling pathway. Reduced functional expression of ribosomal protein S6 as a mechanism involved in sensitizing solid cancers to radiation is also supported in recent findings by Wang et al. [38], that showed genetic inhibitory regulation of this signaling mediator sensitized lung cancer cells to radiation by impairing DNA repair. The observation that down-regulation of MACF1 function reduced ribosomal protein S6 expression is further supported by an investigation by Lee et al. [39], that demonstrated Short-stop, the drosophila homologue of MACF1, interacted with the translational regulator Krasavietz-eIF5C. Taken together the synergistic anti-tumorigenic effects of negatively regulating MACF1 function with radiation in glioblastomas is a culmination of perturbation of the mTOR signaling axis, as indicated by down-regulation of ribosomal protein S6 expression and presumably functional impairment, as well as the co-expression of MACF1 with mTOR and p70s6k in clinical glioblastomas. It can also be postulated that the radiosensitization effects of down-regulating MACF1 are a consequence of a diminished DNA damage repair response, given ribosomal protein S6 and MACF1 expression were positively correlated to regulatory genes associated with this process.
In conclusion, we demonstrated here that inhibiting MACF1 enhances and sensitizes glioblastomas to radiation that extends findings observed with temozolomide and further supports the inhibitory targeting of MACF1 with clinical treatment modalities for these tumors. Furthermore, the mechanistic involvement of MACF1 in intracellular signaling processes has been broadened to include ribosomal protein S6 as part of the mTOR-p70s6k signaling axis that has recently been described as a prognostic indicator related to poor survival of glioblastoma patients [19]. Taken together this investigation has identified MACF1 as a novel radiosensitization target in glioblastomas.
Acknowledgments
Quincy Quick is supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number SC-3GM121178.
References
1. Bernier G, Mathieu M, De Repentigny Y, Vidal SM, Kothary R. Cloning and characterization of mouse ACF7, a novel member of the dystonin subfamily of actin binding proteins. Genomics. 1996; 38:19–29. PMID: 8954775.
2. Ka M, Jung EM, Mueller U, Kim WY. MACF1 regulates the migration of pyramidal neurons via microtubule dynamics and GSK-3 signaling. Dev Biol. 2014; 395:4–18. PMID: 25224226.
3. Hu L, Su P, Li R, et al. Knockdown of microtubule actin crosslinking factor 1 inhibits cell proliferation in MC3T3-E1 osteoblastic cells. BMB Rep. 2015; 48:583–588. PMID: 26277981.
4. Afghani N, Mehta T, Wang J, Tang N, Skalli O, Quick QA. Microtubule actin cross-linking factor 1, a novel target in glioblastoma. Int J Oncol. 2017; 50:310–316. PMID: 27959385.
5. Arai E, Sakamoto H, Ichikawa H, et al. Multilayer-omics analysis of renal cell carcinoma, including the whole exome, methylome and transcriptome. Int J Cancer. 2014; 135:1330–1342. PMID: 24504440.
6. Chang YS, Huang HD, Yeh KT, Chang JG. Identification of novel mutations in endometrial cancer patients by whole-exome sequencing. Int J Oncol. 2017; 50:1778–1784. PMID: 28339086.
7. Chen HJ, Lin CM, Lin CS, Perez-Olle R, Leung CL, Liem RK. The role of microtubule actin cross-linking factor 1 (MACF1) in the Wnt signaling pathway. Genes Dev. 2006; 20:1933–1945. PMID: 16815997.
8. McCord M, Mukouyama YS, Gilbert MR, Jackson S. Targeting WNT signaling for multifaceted glioblastoma therapy. Front Cell Neurosci. 2017; 11:318. PMID: 29081735.
9. Shapiro WR, Green SB, Burger PC, et al. Randomized trial of three chemotherapy regimens and two radiotherapy regimens and two radiotherapy regimens in postoperative treatment of malignant glioma. Brain tumor cooperative group trial 8001. J Neurosurg. 1989; 71:1–9.
10. Khosla D. Concurrent therapy to enhance radiotherapeutic outcomes in glioblastoma. Ann Transl Med. 2016; 4:54. PMID: 26904576.
11. Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009; 10:459–466. PMID: 19269895.
12. Kahn J, Hayman TJ, Jamal M, et al. The mTORC1/mTORC2 inhibitor AZD2014 enhances the radiosensitivity of glioblastoma stem-like cells. Neuro Oncol. 2014; 16:29–37. PMID: 24311635.
13. Gil del Alcazar CR, Hardebeck MC, Mukherjee B, et al. Inhibition of DNA double-strand break repair by the dual PI3K/mTOR inhibitor NVP-BEZ235 as a strategy for radiosensitization of glioblastoma. Clin Cancer Res. 2014; 20:1235–1248. PMID: 24366691.
14. Ahmed SU, Carruthers R, Gilmour L, Yildirim S, Watts C, Chalmers AJ. Selective inhibition of parallel DNA damage response pathways optimizes radiosensitization of glioblastoma stem-like cells. Cancer Res. 2015; 75:4416–4428. PMID: 26282173.
15. King AR, Corso CD, Chen EM, et al. Local DNA repair inhibition for sustained radiosensitization of high-grade gliomas. Mol Cancer Ther. 2017; 16:1456–1469. PMID: 28566437.
16. Shi F, Guo H, Zhang R, et al. The PI3K inhibitor GDC-0941 enhances radiosensitization and reduces chemoresistance to temozolomide in GBM cell lines. Neuroscience. 2017; 346:298–308. PMID: 28147244.
17. Sulzmaier FJ, Ramos JW. RSK isoforms in cancer cell invasion and metastasis. Cancer Res. 2013; 73:6099–6105. PMID: 24097826.
18. Cargnello M, Tcherkezian J, Roux PP. The expanding role of mTOR in cancer cell growth and proliferation. Mutagenesis. 2015; 30:169–176. PMID: 25688110.
19. Ronellenfitsch MW, Zeiner PS, Mittelbronn M, et al. Akt and mTORC1 signaling as predictive biomarkers for the EGFR antibody nimotuzumab in glioblastoma. Acta Neuropathol Commun. 2018; 6:81. PMID: 30129426.
20. Sekhar KR, Benamar M, Venkateswaran A, et al. Targeting nucleophosmin 1 represents a rational strategy for radiation sensitization. Int J Radiat Oncol Biol Phys. 2014; 89:1106–1114. PMID: 25035215.
21. The cBioPortal for Cancer Genomics. The Cancer Genome Atlas. Accessed December 15, 2019. at https://www.cbioportal.org/.
22. Kao GD, Jiang Z, Fernandes AM, Gupta AK, Maity A. Inhibition of phosphatidylinositol-3-OH kinase/Akt signaling impairs DNA repair in glioblastoma cells following ionizing radiation. J Biol Chem. 2007; 282:21206–21212. PMID: 17513297.
23. Kil WJ, Cerna D, Burgan WE, et al. In vitro and in vivo radiosensitization induced by the DNA methylating agent temozolomide. Clin Cancer Res. 2008; 14:931–938. PMID: 18245557.
24. Sheng Y, Xu M, Li C, et al. Nm23-H1 is involved in the repair of ionizing radiation-induced DNA double-strand breaks in the A549 lung cancer cell line. BMC Cancer. 2018; 18:710. PMID: 29970055.
25. Martinou M, Giannopoulou E, Malatara G, Argyriou AA, Kalofonos HP, Kardamakis D. Ionizing radiation affects epidermal growth factor receptor signalling and metalloproteinase secretion in glioma cells. Cancer Genomics Proteomics. 2011; 8:33–38. PMID: 21289335.
26. Contessa JN, Hampton J, Lammering G, et al. Ionizing radiation activates Erb-B receptor dependent Akt and p70 S6 kinase signaling in carcinoma cells. Oncogene. 2002; 21:4032–4041. PMID: 12037685.
27. Kim Y, Kim KH, Lee J, et al. Wnt activation is implicated in glioblastoma radioresistance. Lab Invest. 2012; 92:466–473. PMID: 22083670.
28. Wild-Bode C, Weller M, Rimner A, Dichgans J, Wick W. Sublethal irradiation promotes migration and invasiveness of glioma cells: implications for radiotherapy of human glioblastoma. Cancer Res. 2001; 61:2744–2750. PMID: 11289157.
29. Lammering G, Hewit TH, Valerie K, et al. EGFRvIII-mediated radioresistance through a strong cytoprotective response. Oncogene. 2003; 22:5545–5553. PMID: 12944901.
30. Pelloski CE, Lin E, Zhang L, et al. Prognostic associations of activated mitogen-activated protein kinase and Akt pathways in glioblastoma. Clin Cancer Res. 2006; 12:3935–3941. PMID: 16818690.
31. Li B, Yuan M, Kim IA, Chang CM, Bernhard EJ, Shu HK. Mutant epidermal growth factor receptor displays increased signaling through the phosphatidylinositol-3 kinase/AKT pathway and promotes radioresistance in cells of astrocytic origin. Oncogene. 2004; 23:4594–4602. PMID: 15077177.
32. Liu Q, Nguyen DH, Dong Q, et al. Molecular properties of CD133+ glioblastoma stem cells derived from treatment-refractory recurrent brain tumors. J Neurooncol. 2009; 94:1–19. PMID: 19468690.
33. Shih HA, Betensky RA, Dorfman MV, Louis DN, Loeffler JS, Batchelor TT. Genetic analyses for predictors of radiation response in glioblastoma. Int J Radiat Oncol Biol Phys. 2005; 63:704–710. PMID: 15978739.
34. Barker FG 2nd, Simmons ML, Chang SM, et al. EGFR overexpression and radiation response in glioblastoma multiforme. Int J Radiat Oncol Biol Phys. 2001; 51:410–418. PMID: 11567815.
35. Yang P, Zhang W, Wang Y, et al. IDH mutation and MGMT promoter methylation in glioblastoma: results of a prospective registry. Oncotarget. 2015; 6:40896–40906. PMID: 26503470.
36. Tini P, Nardone V, Pastina P, et al. Patients affected by unmethylated O(6)-methylguanine-DNA methyltransferase glioblastoma undergoing radiochemotherapy may benefit from moderately dose-escalated radiotherapy. Biomed Res Int. 2017; 2017:9461402. PMID: 29159183.
37. Miki S, Imamichi S, Fujimori H, et al. Concomitant administration of radiation with eribulin improves the survival of mice harboring intracerebral glioblastoma. Cancer Sci. 2018; 109:2275–2285. PMID: 29758120.
38. Wang Y, Mei H, Shao Q, Wang J, Lin Z. Association of ribosomal protein S6 kinase 1 with cellular radiosensitivity of non-small lung cancer. Int J Radiat Biol. 2017; 93:581–589. PMID: 28276898.
39. Lee S, Nahm M, Lee M, et al. The F-actin-microtubule crosslinker Shot is a platform for Krasavietz-mediated translational regulation of midline axon repulsion. Development. 2007; 134:1767–1777. PMID: 17409115.
Fig. 1
Correlative expression of MACF1 to patient survival and glioblastoma associated genes. Comparative survival curve analysis of glioblastoma patients with low MACF1 expression (<1.048; n=74) and high MACF1 mRNA expression (>1.048; n=83). MACF1 co-expression with EGFR, PTEN, p53, and CDKN2A.
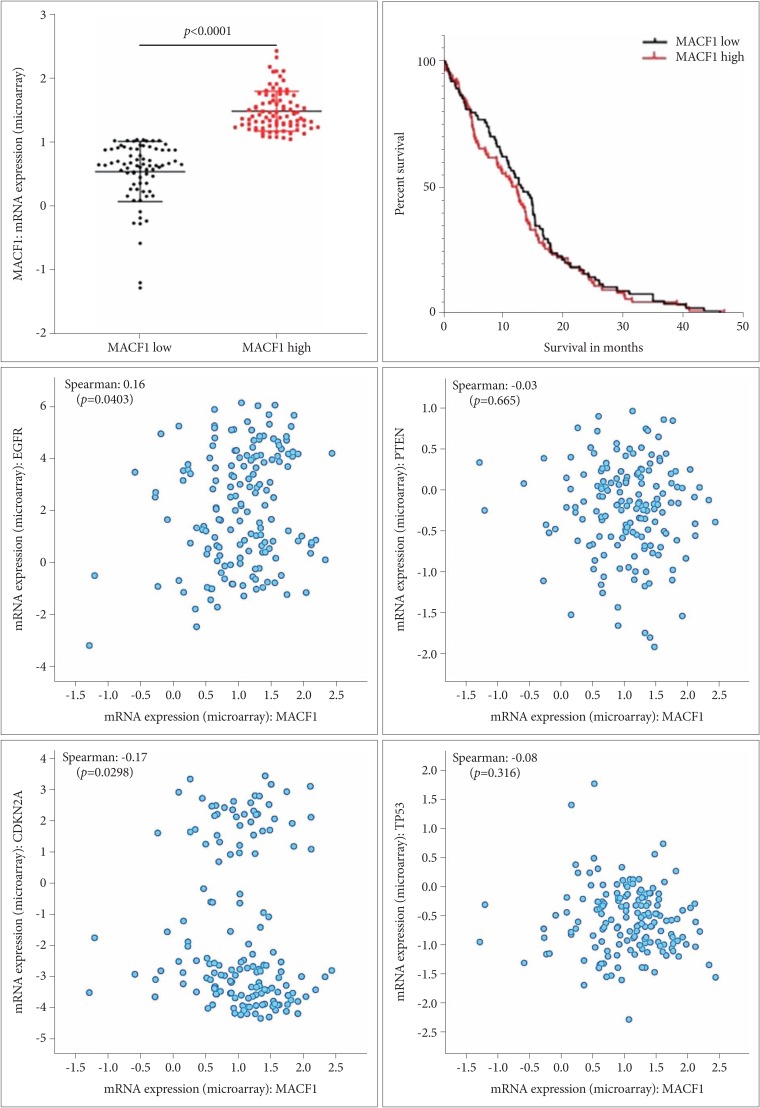
Fig. 2
Down-regulation of MACF1 in genetically silenced glioblastoma cells. U251 glioblastoma cells treated with non-targeting shRNAs (A–C) and shRNAs targeting MACF1 (D–F); A172 glioblastoma cells treated with non-targeting shRNAs (G–I) and shRNAs targeting MACF1 (J–L); MACF1 expression (green); DAPI (blue); cytoplasmic MACF1 expression (arrows). Displayed are immunofluorescent images representative of three separate independent experiments with comparable results. Image J was used to measure the fluorescence intensity from a total of 50 individual cells from three separate independent experiments (n=3) and averaged. Student's t-test was used to determine statistical significance (**p<0.01; error bars: standard error of the means). Scale bar=100 µm; total magnification ×200; RFU: relative fluorescence units.
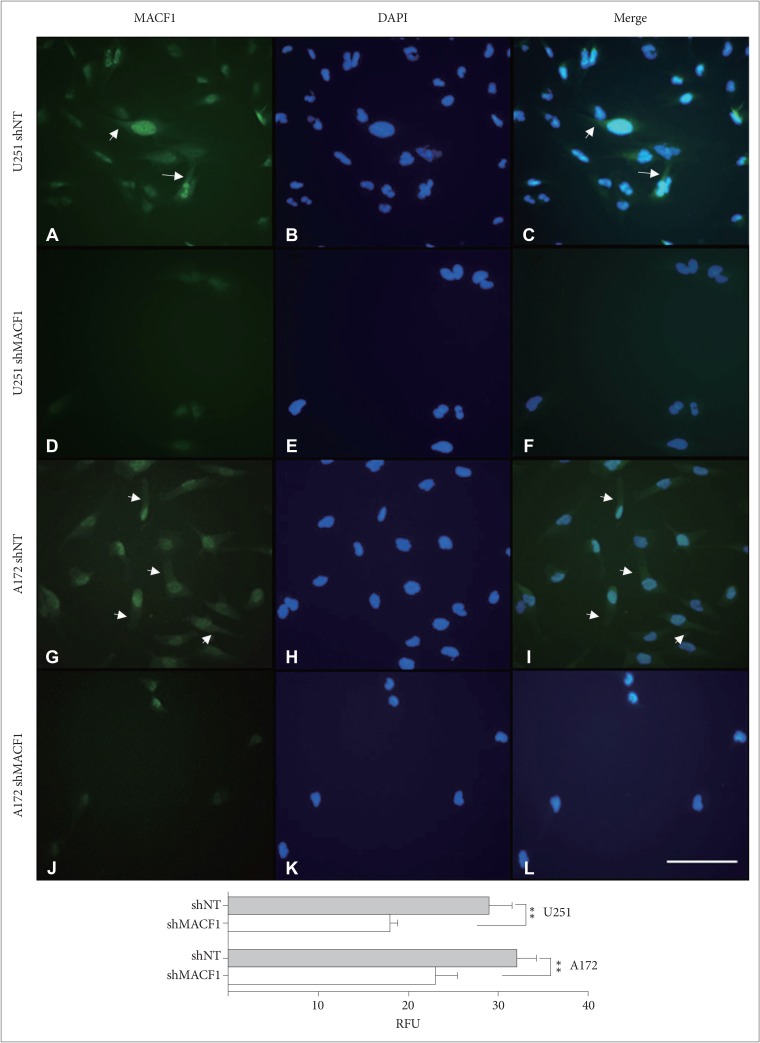
Fig. 3
MACF1 inhibition sensitizes glioblastoma cells to radiation. Impairing MACF1 function reduces cell viability in non-irradiated cells and enhances the efficacy of radiation. Data displayed is of a single experiment performed in duplicate and equivalent to three separate independent experiments performed in duplicate with comparable outcomes. ANOVA analysis followed by Tukey's post-hoc analysis revealed statistical significant differences between means of experimental conditions (*p<0.05; **p<0.01; error bars: standard error of the means).
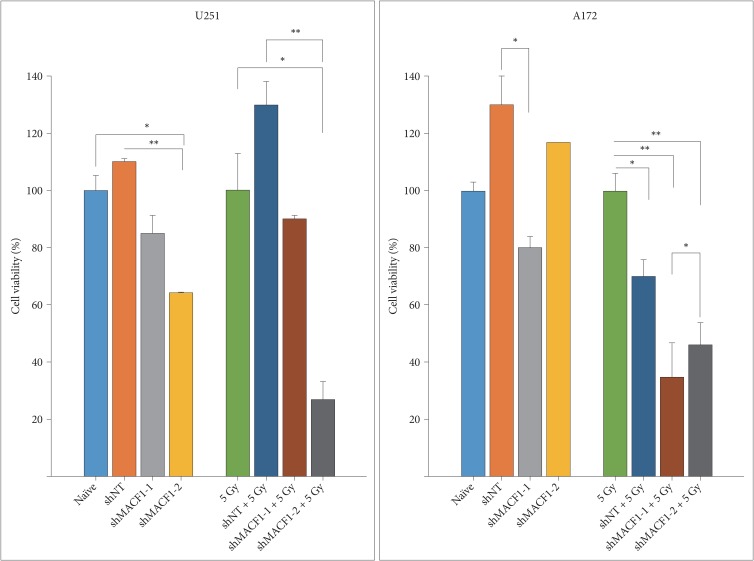
Fig. 4
Radiation and suppression of MACF1 impairs glioblastoma cell migration. U251 (A–D) and A172 (E–H) cells treated with shMACF1 and irradiated with a single 5 Gy dose. Displayed are stained migrating cells in Boyden chambers and data from a single experiment performed in duplicate representative of at least three separate independent experiments performed in duplicate with comparable results 24 hours after radiation exposure. ANOVA analysis followed by Tukey's post-hoc analysis revealed statistical significant differences between means of experimental conditions (*p<0.05; **p<0.01; error bars: standard error of the means). Scale bar=200 µm; stained with crystal violet; total magnification ×100.
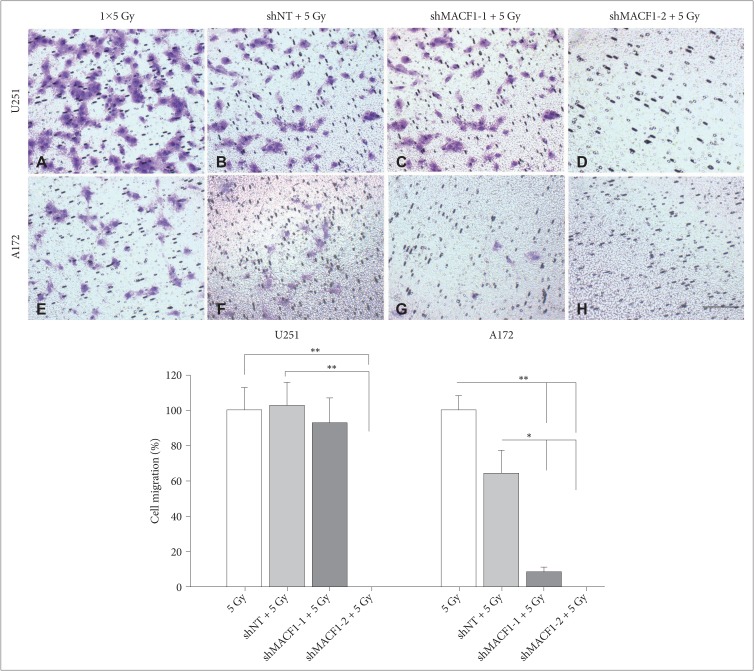
Fig. 5
Evaluation of translational regulatory proteins as mechanistic components of the radiosensitization effects of downregulating MACF1. A, B: Positive correlative expression of MACF1 with ribosomal protein S6 regulators MTOR and RPS6KB1 in clinical glioblastoma patients. C: Silencing MACF1 combined with radiation decreases p-ribosomal protein S6 expression. Immunofluorescence images displayed are from a single experiment in U251 glioblastoma cells at 24 hours and representative of three separate experiments that showed comparable results. Image J was used to measure the fluorescence intensity from a total of 50 individual cells from three separate independent experiments (n=3) and averaged. ANOVA analysis followed by Tukey's post-hoc analysis revealed statistical significant differences between means of experimental conditions (****p<0.0001; error bars: standard error of the means). Scale bar=80 µm; total magnification ×400. RFU: relative fluorescence units.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download