Abstract
Digital pathology is revolutionizing pathology. The introduction of digital pathology made it possible to comprehensively change the pathology diagnosis workflow, apply and develop pathological artificial intelligence (AI) models, generate pathological big data, and perform telepathology. AI algorithms, including machine learning and deep learning, are used for the detection, segmentation, registration, processing, and classification of digitized pathological images. Pathological AI algorithms can be helpfully utilized for diagnostic screening, morphometric analysis of biomarkers, the discovery of new meanings of prognosis and therapeutic response in pathological images, and improvement of diagnostic efficiency. In order to develop a successful pathological AI model, it is necessary to consider the selection of a suitable type of image for a subject, utilization of big data repositories, the setting of an effective annotation strategy, image standardization, and color normalization. This review will elaborate on the advantages and perspectives of digital pathology, AI-based approaches, the applications in pathology, and considerations and challenges in the development of pathological AI models.
Nowadays, pathology has undergone dramatic transformations due to digitization and the introduction of new computing technologies, in line with other medical fields. Digital pathology is ending the era of optical microscopy, which has lasted over 100 years, and is ushering in a new era in which digitized whole slide images (WSIs) are utilized for pathology diagnoses. The advent of digital pathology systems has facilitated expedited clinical pathological workflows, a new model of education, and the development of practically actionable image analysis tools and investigation techniques [1].
Pathological data analysis using artificial intelligence (AI) is considered an essential approach in the age of precision oncology [2]. In cancer, the complexity of genomic and proteomic alterations and cellular characteristics and interactions between tumor cells and their microenvironment can influence the progression of the disease and affect therapeutic response. Pathological assessment of these alterations requires the simultaneous interrogation of multiple features in a highly sensitive and accurate approach. In addition, well-validated and reproducible assessment tools are needed for clinical decision-making. Although the extraction of multiple visual and morphometric AI-based features from the pathological WSI is still limited due to slide quality problems and tumor heterogeneity, it can help to overcome the limitations of the pathologist’s subjective visual and semi-quantitative evaluation and comprehensively analyze complex tissue architectures.
This review will elaborate on the advantages and perspectives of digital pathology, AI-based approaches, the applications in pathology, and considerations and challenges in the development of pathological AI models.
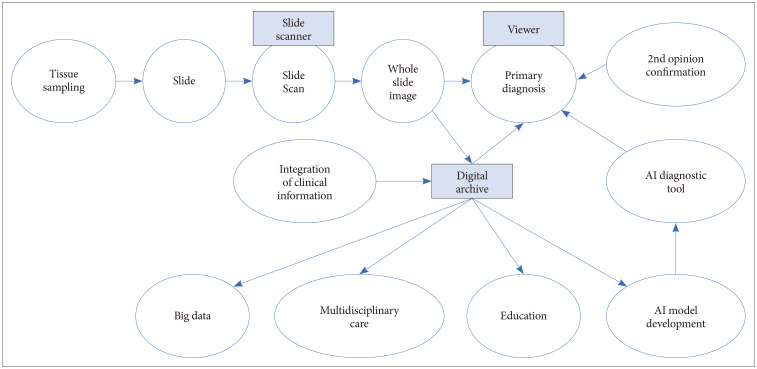
Digital pathology was initially referred to as the process of digitizing WSIs using advanced slide scanning technology, and now is a generic term that includes AI-based approaches for detection, segmentation, diagnosis, and analysis of digitalized images [2]. Digital pathology drives changes across pathology, including pathological diagnostic workflows, telepathology, the adoption of various AI-based diagnostic algorithms in routine diagnosis, pathology education, and big data generation and management [3]. A digital pathology system allows a pathologist to observe digitized images on a computer monitor rather than examining tissue glass slides using a microscope in the diagnostic process (Fig. 1) [4]. In detail, a patient tissue specimens obtained through biopsy or excision is fixed in formalin to prepare a paraffin tissue block. The paraffin block is sliced into thin sections off 3–4 µm thickness, and the thin section are placed onto a glass slide. These processes are the same as the existing slide preparing method. In a digital pathology system, the tissue glass slides are digitized using a slide scanner after the tissue slide preparation is completed. The slide scanner, equipped with a lens with a magnification of ×20 or ×40, scans in a line or patch method, stitches scanned images together, and then makes WSIs. As with examining tissue slides under a microscope, pathologists can observe all areas of the WSI at various magnifications. A WSI is a gigapixel, ultrahigh-resolution image and is approximately 1 GB in size when a 1.5 cm×1.5 cm tissue section is scanned. A digital pathology system should be equipped with a slide scanner and an image viewer capable of viewing WSI. To operate the system properly in a hospital, very fast communication network, advanced slide compression technology, the latest streaming technology, sufficient storage server, and full integration with the other medical systems such as electronic medical systems (EMR) or picture archiving and communication systems for radiology are essential.
Recently, primary diagnosis in digital pathology systems has been approved and put into practical use in Korea as well as the United States, the European Union, and Japan [3]. Many studies have compared the diagnostic accuracy of primary diagnosis in digital pathology systems with conventional microscopic diagnostics [567]. Most of the validation studies have demonstrated that routine diagnosis with a digital pathology system is not inferior to conventional diagnostics, and a recent meta-analysis study demonstrated equivalent performance of digital pathology in comparison with light microscopy for routine diagnosis [8]. Mukhopadhyay et al. [5] evaluated the diagnostic performance of digitalized images compared to microscopic images on specimens from 1,992 patients with different tumor types diagnosed by 16 surgical pathologists. Routine diagnostic performance with digitalized WSIs was not inferior to that achieved with light microscopy-based approaches (with a major discordance rate from the reference standard of 4.9% for WSI and 4.6% for microscopy). Through internet networks, digital pathology enables real-time mutual consultation about the case with many pathological experts at other institutions and in different countries [9]. In addition, pathological diagnosis can be made without time and space limitations in the digital pathology system. Pathological diagnosis requires a detailed review of all medical data and past pathological data of the patient, and digital pathology enables the integrated analysis of data-based medical data. One of the important advantages of digital pathology is that pathological AI-based models can be easily used during the diagnostic process and collection and management of pathologic big data [10]. Pathological big data can be used for learning various AI diagnostic models needed for pathological diagnosis and can be used as various educational materials [11].
Digital pathology is still evolving and has many challenges to overcome. Currently, digital pathology is being introduced in several hospitals around the world, but there are only a few so far. There are relatively a few reports based on digital pathology experience to completely replace light microscopy for routine diagnostic purposes [1213]. The digital pathology system is supplied by several commercial companies, but the technical compatibility between them is an issue. The initial investment for having a digital pathology system is high because a slide scanner, viewer, and storage server are required. A UK pathology laboratory focus group published that they identified benefits (e.g., collaboration, teaching, and cost-savings) of the implementation of digital pathology [14]. It is important to integrate digital pathology systems to make smooth use of digital pathology systems. This requires a lot of time, manpower, cost, and collaboration with existing system operators. In addition, the level of operation of the digital pathology system may be limited depending on the level of computerization of the pathology order communication system or EMR system [3].
As qualified pathological big data have been accumulated due to the introduction of digital pathology to medical institutions with the development of scanning technology, pathologists can explore various unresolved problems in pathology by collaborating with technical experts, including data scientists, computational engineers, and image physicists [15]. Computer vision tasks include methods for acquiring, processing, analyzing, and understanding digital images, and extraction of high-dimensional data from the real world to produce numerical or symbolic information [16]. AI and machine learning (ML) technologies are ideal for examining complex data sets in an exhaustive manner and have been deployed to extract features, patterns, and information from histopathological images [4]. Deep learning (DL) is a concept belonging to ML and consists of computer algorithms that learn by themselves as they perform tasks and has strengths in extracting features from medical images, speech recognition, and natural language processing [17]. In pathology, DL provides an opportunity to interrogate pathological images at a deeper level, and DL algorithms are used for detection, segmentation, registration, processing, and classification of digitized WSI [18].
Pathological AI models can be used for various purposes, including automatic detection, localization, quantification of histological parameters and structural changes, and disease diagnosis [18]. These models support pathologists in diagnosing disease by analyzing the characteristics of tumor cells, structural changes of lesions, and expression patterns of biomarkers. In addition to histopathological image analysis, AI models can comprehensively investigate various medical metadata (e.g., clinical, genetic, radiological, laboratory data, etc.) coupled with histopathological image data [4]. Through “omics” analysis, AI models may predict disease aggressiveness, patient outcomes, and therapeutic response. These new capabilities of AI models can extract useful information in ways that were not possible in conventional microscopic diagnostics in the context of direct human cognitive and visual assessment.
The increase in quantitative and qualitative demands for pathological diagnosis for the realization of precision medicine is further promoting the application of AI models in pathological diagnosis. While applying DL approaches to pathology in the early days, AI models that mimic pathologists were mainly developed with the goal of reducing the burden of pathologists or increasing the accuracy of pathological diagnosis [19]. For example, prostate cancer detection and localization models for needle biopsy specimen have been approved for clinical practice [20]. Using DL algorithm, biomarker morphometric analysis models to determine patients eligible for targeted therapy, for Ki67, c-erbB2, estrogen receptor, and progesterone receptor expressions, have been developed by the Asan Medical Center in collaboration with Vuno Inc. and approved by the Ministry of Food and Drug Safety in Korea. Prediction models of microsatellite instability in colon cancer and non-small cell lung cancer that have EGFR or KRAS gene mutations are also being actively developed and validated [2122]. These predictive models are expected to be used for screening purposes in the pathological diagnosis process, and they aim to minimize unnecessary molecular genetic testing and reduce the waste of time and medical expenses. With the accumulation of pathological big data, AI models for rare diseases or differential diagnosis, which are one of the most unmet needs, have recently been proposed [23].
Pathological diagnosis is a confirmative diagnosis of a disease, and pathological diagnosis plays an absolutely important role in the determination of the treatment plan of patients in neurooncology. The recent trend in WHO central nervous system (CNS) tumors classification has highly emphasized the importance of genetic mutations in addition to the conventional morphological and immunophenotypical characteristics [24]. Many types of targeted therapeutic and immunotherapeutic agents are being developed for CNS tumors. In line with the development of therapeutic agents, various biomarkers are being excavated to select the most effective therapeutic agents and to predict and evaluate the responsiveness of treatment [25]. For these reasons, currently, the evaluation of morphological, immunophenotypical, and genetic biomarkers associated with an accurate diagnosis and precision medicine using tumor tissue in pathological diagnosis is essential. To evaluate the expression of biomarkers and the aberration of genes, many kinds of ancillary tests, such as immunohistochemical staining, fluorescent in situ hybridization analysis, Sanger’s sequencing, next-generation sequencing, and PCR-based DNA methylation analysis, are required. To perform these tests, it takes a lot of money and a long time, and a considerable amount of tissue is consumed. In order to manage the patient at the appropriate time, it may be very difficult to carry out all the tests. Studies have reported that AI-based algorithms can predict the expression of biomarkers and genetic mutations of tumors by evaluating only the morphological characteristics of hematoxylin and eosin (H&E)-stained slides that are basically produced for pathological diagnosis [2122262728]. Studies have also shown that novel biomarkers can be discovered by utilizing AI models in various types of cancers [293031]. Therefore, for the patient’s appropriate care, it is important to use AI in neurooncology to screen for pathological classification or to predict the characteristics of cancer.
Recently, pathological AI models are being actively developed in the field of neurooncology, and they mainly target gliomas. In detail, pathological AI models being developed focus on the classification of gliomas, the mutation status of cancer-related genes, predicting patients’ prognosis, discovering new histological implicating features of tumors affecting cancer behaviors, and analyzing tumor microenvironment [2728323334]. Most of the research uses H&E-stained slides, immunohistochemical stained slides, and, in some cases, genetic analysis information. Jin et al. [27] developed a convolutional neural network model for glioma classification. This model classified five major histological subtypes of glioma, which were oligodendroglioma, anaplastic oligodendroglioma, astrocytoma, anaplastic astrocytoma, glioblastoma, and nontumor, with high patient-level accuracy of 87.5% using H&E-stained slide, IDH mutation, and 1p/19q deletion. Pathological AI models were proposed for glioma grading and for estimating IDH mutation status in low-grade glioma and for predicting the patient’s prognosis on H&E-stained slides [3233]. Zadeh Shirazi et al. [34] defined the tumor cell-perivascular niche and spatial associated genetic signatures using a semantic segmentation model and demonstrated that they were associated with poor survival in glioblastoma.
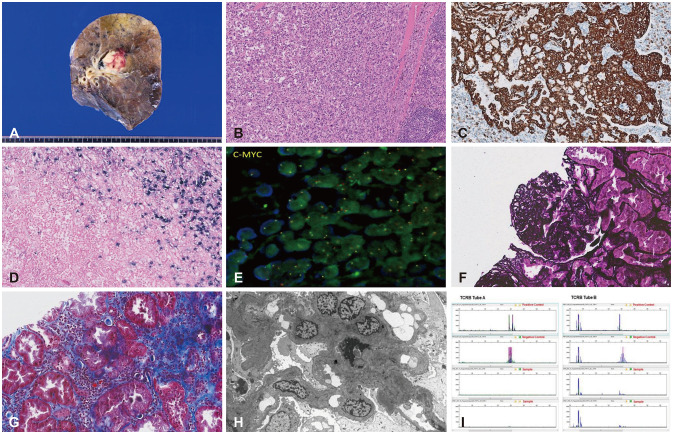
Pathological AI algorithms can be helpfully utilized for diagnostic screening, morphometric analysis of biomarkers, the discovery of new meanings of prognosis and therapeutic response in pathological images, and improvement of diagnostic efficiency. In the case of determining a subject for a pathological AI model, it is necessary to collect relevant data. Histopathological images are basically stained with H&E and most of all surgical pathology diagnoses are made based on H&E-stained slides. Now, the majority of pathological AI models have been developed using H&E-stained WSI. Most of the pathological data is image data, but various types of pathological images are created depending on the staining methods and the examination methods. Pathology produces various types of images to observe macroscopic, histological, and cellular-level morphological changes, increases or decreases in copy number, translocation, and mutation of DNA, protein expression patterns, mRNA expression patterns, microorganisms, and ultrastructural morphological changes (Fig. 2). Since WSI shows its own histological characteristics according to the applied staining methods and is used for different purposes for pathological diagnosis, a pathological AI model can be developed using one type of image data but can also be developed using several different types of images depending on the subject. Therefore, it is important to determine and collect the kind of pathological image data that are best suited to the topic of the AI model to be developed.
Because big data is essential for AI model development, data repositories are being actively built by medical organizations/associations and medical institutions. TCGA is one of the most representative data repositories in the field of cancer, and it contains clinical data and genetic data along with pathological WSIs, which have been widely used for learning and validation of pathological AI model development [263536]. There are currently several data repositories in the field of renal pathology, such as NEPTUNE, CureGN, and FOrNE [4]. Many medical institutions, led by developed countries, have been able to actively collect pathological big data by implementing digital pathology systems. Until recently, one of the biggest difficulties in developing pathological AI models was the lack of pathological big data, but it is expected that many of these difficulties will be resolved within the next few years. Moreover, as transfer learning and synthetic data-related technologies have recently started to be applied to pathology, it is expected that the problem of data shortage can be overcome in a shorter period [1037].
A pathological examination is performed very rarely compared to other medical examinations such as radiologic and laboratory examinations. Currently, pathological big data is being actively generated, but there is very little data available to develop a pathological AI algorithm that can handle the complex characteristics of pathological image data and subjects that are very difficult to solve but, at the same time, require accuracy of output. So, annotations are indispensable for pathological AI model development. Pathological image annotation is too difficult for anyone other than pathologists to perform. In addition, even when performed by a pathologist, it can be time-consuming, taking up to several hours per case, and in some cases, it must be performed by a highly specialized pathologist. Therefore, when developing a pathological AI algorithm, generating annotation data rather than collecting digitized images often becomes the bottleneck of development. In the case of using fully supervised learning to develop a pathological AI model, a strategic approach that can minimize annotation burden is required, and when a relatively large amount of data can be collected, it is necessary to consider weakly supervised learning methods. In the case of a cancer diagnosis model for prostate cancer needle biopsy diagnosis, multiple instance learning, one of the weakly supervised learning methods, was applied to develop an AI model with high accuracy through simple labeling without detailed annotation of images [38]. Active learning is a kind of semi-supervised learning in which a learning algorithm is allowed to interactively ask a user to provide new labels for samples [39]. Active learning suggests that a learning algorithm can perform better with less data than traditional methods if it can choose the data it wants to learn from. In pathology, active learning has been successfully applied to classifiers to support quality control in nucleus segmentation [40]. The main hypothesis in transfer learning is that if a previously well-trained algorithm is applied to a different but related problem, it can perform better than traditional methods with substantially less data for training, and transfer learning frameworks are also actively used in pathology. Annotations are available on platforms that can handle pathological images. Annotation of WSIs in various formats can be performed through open platforms such as Automated Slide Analysis Platform (ASAP) (https://computationalpathologygroup.github.io/ASAP/) or Quantitative Pathology & Bioimage Analysis (QuPath) (https://qupath.github.io/) in addition to commercial software. Of note, these platforms can perform image overlays or use open-source pathological image analysis tools.
WSIs are not standardized yet, unlike radiological data that are standardized as digital image and communication model (DICOM, www.dicomstandard.org) format. Slide scanner manufacturers use the most appropriate image format for WSIs, such as .tif, .svs, .ndpi, .mrxs, bif., and .isyntax, produced by their scanners. The lack of a standard image format is a major obstacle not only to the routine diagnosis of digital pathology but also to the development of pathological AI models. For routine diagnosis in digital pathology, each medical institution must be equipped with an image viewer that can operate all image formats or a digital pathology system that can convert to an image format suitable for their own viewer. Even when developing an AI model, it is necessary to be able to read the collected WSIs. A library that can read various image formats can be provided by open sources such as OpenSlide (www.openslide.org). Efforts to establish a pathological DICOM standard format have been made in recent years [41]. WG-26 is a US- and European-led pathology working group within DICOM. The scope of WG-26 is to support and develop the DICOM standard, allowing the pathology domain to process WSIs and macros to produce, store, and communicate. They try to open up those systems for interoperability using global standards to create workflows as a part of the overall healthcare process. WSIs show different image characteristics, even on H&E-stained slides, depending on the difference that occurs in the slide production process and the brand or model of a slide scanner. Differences in image characteristics affect the learning of AI algorithms and may cause performance degradation when the developed AI model is applied. Color normalization approaches have been made to compensate for these variations [42]. Stain-color deconvolution and template color matching have been used for color normalization of histopathological images. The stain-color deconvolution method considers prior knowledge of the reference staining vector for all dyes and splits an input RGB image into three staining channels, each representing the actual staining color [43]. Template color matching achieves color correction by choosing an appropriate source image and applying its characteristics to another image [44]. Generative adversarial networks are also used for histopathological images [45].
Digital pathology is currently leading a comprehensive shift in pathology. The implementation of digital pathology impacts pathological diagnostic workflows, telepathology, the adoption of various AI-based diagnostic algorithms in routine diagnosis, pathology education, and big data generation and management. Pathological AI algorithms can be helpfully utilized for diagnostic screening, morphometric analysis of biomarkers, the discovery of new meanings of prognosis and therapeutic response in pathological images, and improvement of diagnostic efficiency. In order to develop a successful pathological AI model, it is necessary to consider the selection of a suitable type of image for a subject, the utilization of big data repositories, the setting of an effective annotation strategy, image standardization, and color normalization.
Availability of Data and Material
Data sharing not applicable to this article as no datasets were generated or analyzed during the study.
References
1. Nam S, Chong Y, Jung CK, Kwak TY, Lee JY, Park J, et al. Introduction to digital pathology and computer-aided pathology. J Pathol Transl Med. 2020; 54:125–134. PMID: 32045965.
2. Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019; 16:703–715. PMID: 31399699.
3. Jahn SW, Plass M, Moinfar F. Digital pathology: advantages, limitations and emerging perspectives. J Clin Med. 2020; 9:3697.
4. Barisoni L, Lafata KJ, Hewitt SM, Madabhushi A, Balis UGJ. Digital pathology and computational image analysis in nephropathology. Nat Rev Nephrol. 2020; 16:669–685. PMID: 32848206.
5. Mukhopadhyay S, Feldman MD, Abels E, Ashfaq R, Beltaifa S, Cacciabeve NG, et al. Whole slide imaging versus microscopy for primary diagnosis in surgical pathology: a multicenter blinded randomized noninferiority study of 1992 cases (pivotal study). Am J Surg Pathol. 2018; 42:39–52. PMID: 28961557.
6. Goacher E, Randell R, Williams B, Treanor D. The diagnostic concordance of whole slide imaging and light microscopy: a systematic review. Arch Pathol Lab Med. 2017; 141:151–161. PMID: 27399211.
7. Snead DR, Tsang YW, Meskiri A, Kimani PK, Crossman R, Rajpoot NM, et al. Validation of digital pathology imaging for primary histopathological diagnosis. Histopathology. 2016; 68:1063–1072. PMID: 26409165.
8. Azam AS, Miligy IM, Kimani PK, Maqbool H, Hewitt K, Rajpoot NM, et al. Diagnostic concordance and discordance in digital pathology: a systematic review and meta-analysis. J Clin Pathol. 2021; 74:448–455. PMID: 32934103.
9. van Diest PJ, Huisman A, van Ekris J, Meijer J, Willems S, Hofhuis H, et al. Pathology image exchange: the Dutch digital pathology platform for exchange of whole-slide images for efficient teleconsultation, telerevision, and virtual expert panels. JCO Clin Cancer Inform. 2019; 3:1–7.
10. Asa SL, Bodén AC, Treanor D, Jarkman S, Lundström C, Pantanowitz L. 2020 vision of digital pathology in action. J Pathol Inform. 2019; 10:27. PMID: 31516758.
11. Boyce BF. Whole slide imaging: uses and limitations for surgical pathology and teaching. Biotech Histochem. 2015; 90:321–330. PMID: 25901738.
12. Alami H, Fortin JP, Gagnon MP, Pollender H, Têtu B, Tanguay F. The challenges of a complex and innovative telehealth project: a qualitative evaluation of the Eastern Quebec Telepathology Network. Int J Health Policy Manag. 2018; 7:421–432. PMID: 29764106.
13. Evans AJ, Salama ME, Henricks WH, Pantanowitz L. Implementation of whole slide imaging for clinical purposes: issues to consider from the perspective of early adopters. Arch Pathol Lab Med. 2017; 141:944–959. PMID: 28440660.
14. Turnquist C, Roberts-Gant S, Hemsworth H, White K, Browning L, Rees G, et al. On the edge of a digital pathology transformation: views from a cellular pathology laboratory focus group. J Pathol Inform. 2019; 10:37. PMID: 31897354.
15. Abels E, Pantanowitz L, Aeffner F, Zarella MD, van der Laak J, Bui MM, et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J Pathol. 2019; 249:286–294. PMID: 31355445.
16. Klette R. Concise computer vision. London: Springer;2014.
17. Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge: MIT Press;2016.
18. Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J Pathol Inform. 2016; 7:29. PMID: 27563488.
19. Holten-Rossing H, Talman MM, Jylling AMB, Laenkholm AV, Kristensson M, Vainer B. Application of automated image analysis reduces the workload of manual screening of sentinel lymph node biopsies in breast cancer. Histopathology. 2017; 71:866–873. PMID: 28677240.
20. Stephens K. FDA authorizes prostate AI software [Internet]. Overland Park: AXIS Imaging News;2021. 09. 22. Accessed Dec 12, 2021. at https://axisimagingnews.com/radiology-products/radiology-software/ai-machine-learning/fda-authorizes-prostate-ai-software.
21. Echle A, Grabsch HI, Quirke P, van den Brandt PA, West NP, Hutchins GGA, et al. Clinical-grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology. 2020; 159:1406–1416.e11. PMID: 32562722.
22. Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018; 24:1559–1567. PMID: 30224757.
23. Lu MY, Zhao M, Shady M, Lipkova J, Chen TY, Williamson DFK, et al. Deep learning-based computational pathology predicts origins for cancers of unknown primary. arXiv [Preprint]. 2020; cited 2021 Dec 20. Available at: . DOI: 10.48550/arXiv.2006.13932.
24. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021; 23:1231–1251. PMID: 34185076.
25. Jain KK. A critical overview of targeted therapies for glioblastoma. Front Oncol. 2018; 8:419. PMID: 30374421.
26. Zhou C, Jin Y, Chen Y, Huang S, Huang R, Wang Y, et al. Histopathology classification and localization of colorectal cancer using global labels by weakly supervised deep learning. Comput Med Imaging Graph. 2021; 88:101861. PMID: 33497891.
27. Jin L, Shi F, Chun Q, Chen H, Ma Y, Wu S, et al. Artificial intelligence neuropathologist for glioma classification using deep learning on hematoxylin and eosin stained slide images and molecular markers. Neuro Oncol. 2021; 23:44–52. PMID: 32663285.
28. McAvoy M, Prieto PC, Kaczmarzyk JR, Fernández IS, McNulty J, Smith T, et al. Classification of glioblastoma versus primary central nervous system lymphoma using convolutional neural networks. Sci Rep. 2021; 11:15219. PMID: 34312463.
29. Harder N, Schönmeyer R, Nekolla K, Meier A, Brieu N, Vanegas C, et al. Automatic discovery of image-based signatures for ipilimumab response prediction in malignant melanoma. Sci Rep. 2019; 9:7449. PMID: 31092853.
30. Brockmoeller S, Echle A, Ghaffari Laleh N, Eiholm S, Malmstrøm ML, Plato Kuhlmann T, et al. Deep learning identifies inflamed fat as a risk factor for lymph node metastasis in early colorectal cancer. J Pathol. 2022; 256:269–281. PMID: 34738636.
31. Yao J, Wang S, Zhu X, Huang J. Imaging biomarker discovery for lung cancer survival prediction. In : Proceedings of the 19th International Conference on Medical Image Computing and Computer-Assisted Intervention; 2016 Oct 17-21; Athens, Greece. Springer;2016. p. 649–657.
32. Jiang S, Zanazzi GJ, Hassanpour S. Predicting prognosis and IDH mutation status for patients with lower-grade gliomas using whole slide images. Sci Rep. 2021; 11:16849. PMID: 34413349.
33. Rathore S, Niazi T, Iftikhar MA, Chaddad A. Glioma grading via analysis of digital pathology images using machine learning. Cancers (Basel). 2020; 12:578.
34. Zadeh Shirazi A, McDonnell MD, Fornaciari E, Bagherian NS, Scheer KG, Samuel MS, et al. A deep convolutional neural network for segmentation of whole-slide pathology images identifies novel tumour cell-perivascular niche interactions that are associated with poor survival in glioblastoma. Br J Cancer. 2021; 125:337–350. PMID: 33927352.
35. Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015; 19:68–77.
36. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018; 173:400–416.e11. PMID: 29625055.
37. Thorstenson S, Molin J, Lundström C. Implementation of large-scale routine diagnostics using whole slide imaging in Sweden: digital pathology experiences 2006-2013. J Pathol Inform. 2014; 5:14. PMID: 24843825.
38. Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019; 25:1301–1309. PMID: 31308507.
39. Settles B. Active learning literature survey. Computer sciences technical report. Madison: University of Wisconsin - Madison;2009. Report no. 1648.
40. Wen S, Kurc TM, Hou L, Saltz JH, Gupta RR, Batiste R, et al. Comparison of different classifiers with active learning to support quality control in nucleus segmentation in pathology images. AMIA Jt Summits Transl Sci Proc. 2018; 2018:227–236.
41. Herrmann MD, Clunie DA, Fedorov A, Doyle SW, Pieper S, Klepeis V, et al. Implementing the DICOM standard for digital pathology. J Pathol Inform. 2018; 9:37. PMID: 30533276.
42. Vahadane A, Peng T, Sethi A, Albarqouni S, Wang L, Baust M, et al. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imaging. 2016; 35:1962–1971. PMID: 27164577.
43. Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001; 23:291–299. PMID: 11531144.
44. Reinhard E, Adhikhmin M, Gooch B, Shirley P. Color transfer between images. IEEE Comput Graph Appl. 2001; 21:34–41.
45. Zhou N, Cai D, Han X, Yao J. Enhanced cycle-consistent generative adversarial network for color normalization of H&E stained images. In : Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019; 2019 Oct 13-17; Shenzhen, China. Springer;2019. p. 694–702.
Fig. 2
Various types of pathological images. A: Gross image. B: Hematoxylin and eosin (H&E) stain. C: 3,3'-Diaminobenzidine (DAB) staining immunohistochemistry. D: In situ hybridization. E: Fluorescent in situ hybridization. F: Periodic acid methenamine silver stain. G: Masson's trichrome stain. H: Transmission electron microscopic image. I: BIOMED-2. PCR assay for T-cell monoclonality image.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download