Abstract
Glioblastoma multiforme (GBM), a high-grade astrocytic brain tumor, has highly aggressive and heterogeneous phenotypes with active cellular invasion, angiogenesis, and immune system modulation in the tumor microenvironment driven by complex oncogenic mutations. This abnormal disease progression could be attributed to extracellular vesicles (EVs) containing diverse bioactive molecules, including proteins, genetic materials, lipids, and metabolites. Importantly, GBM-related EVs have emerged as key mediators in cancer progression, acting as carriers for the transfer of oncogenic proteins such as epidermal growth factor receptor variant III (EGFRvIII) and genetic materials (DNA and RNA). Remarkably, recent progress in EV analysis has enabled its purification with high confidence by estimating the purity level of isolated EVs. Thus, mass spectrometry-based proteomic analysis could generate highly reliable vesicular proteomes. Glioblastoma EV proteome studies have revealed the specific increase in vesicular protein cargo due to their oncogenic transformation, and these EV proteins are closely associated with cancer invasion. Moreover, their proteomic data reflects the molecular alterations that occur in parental GBM and provides potent diagnostic information in a minimally invasive manner in liquid biopsy. Thus, proteomic analysis of GBM EVs could provide an increased understanding of their biological properties and activity in the GBM microenvironment, and provide significant implications for advanced approaches in the diagnosis of these intractable tumors.
Extracellular vesicles (EVs) are lipid bilayer vehicles that range from 30 nm to 1 µm in size and are secreted by most cells [1]. Although they are composed of selected proteins, genetic materials (RNA and DNA), lipids, and metabolites representing the state of the cells they are derived from [2], EVs are highly heterogeneous depending on their size and molecular composition [3]. Traditionally, EVs are classified as exosomes and ectosomes based on their mechanism of biogenesis [4]. Exosomes are smaller, measuring 30–150 nm, and were first observed in endocytic transferrin receptor recycling during reticulocyte maturation [56]. Later studies have revealed the detailed molecular mechanism of exosome biogenesis with the formation of multivesicular bodies (MVB) by either the endosomal sorting complexes required for transport (ESCRT) machinery [7] or the sphingomyelinase-ceramide pathway [8]. These MVB fuse to the plasma membrane [9], resulting in the release of intraluminal vesicles into extracellular space and are referred to as exosomes. Ectosomes are relatively large EV subtypes, ranging from 100 nm to 1 µm. These are directly shed from the plasma membrane which is regulated by actin cytoskeleton rearrangement for fission events [10]. It is well-known that the shedding of ectosomes is specifically regulated by ADP-ribosylation factor 6 (ARF6) [11] and a recent report suggested that annexin A1 (ANXA1) is a specific marker protein in the shedding of ectosomes [12]. EVs cannot be classified following their release because their size, molecular composition, and charge are quite similar and overlap each other [13]. Thus, the International Society for Extracellular Vesicles recommends the use of the umbrella term EVs to cover all secreted vesicle subtypes [13].
Glioblastoma multiforme (GBM) is a common aggressive astrocytic brain tumor with severe morbidity and an average five-year survival rate of 6.8% [14]. Its poor prognosis is associated with rapid cellular proliferation and extensive angiogenesis, accompanied by therapeutic resistance [1516]. Moreover, the GBM therapeutic bottleneck remains due to recurrence after surgical resection and non-responsiveness to radiation therapy or chemotherapy drugs such as temozolomide (TMZ) [1417]. While most cases of GBM show morphological similarities, these tumors are also characterized by molecular differences classified as proneural, classical, and mesenchymal subtypes [14]. Various oncogenic mutations are closely associated with these subtypes. Platelet-derived growth factor receptor A (PDGFRA) or cyclin-dependent kinase 4 (CDK4) amplification favors the formation of oligodendrocyte-like [18] or neural progenitor-like cells [19], respectively. Deletion or mutation of neurofibromin 1 (NF1) is associated with the formation of mesenchymal-like cells in the mesenchymal GBM subtype and epidermal growth factor receptor (EGFR) amplification/mutation drives the generation of astrocytic-like cells in the classical GBM subtype [20]. Approximately 40%–70% of GBMs exhibit abnormal EGFR expression, including EGFR gene amplification and mutation known as EGFR variant III (EGFRvIII) [212223], which has a truncated ligand-binding domain that enables constitutive ligand-independent transformation and prevents its downregulation via endocytosis [2425].
These oncogenic transformations of GBM cells affect their cancerous properties, including abnormal proliferation and invasiveness, and activate the mechanism of EV biogenesis accompanied by deregulation of intercellular pathways in a non-cell-autonomous manner. Importantly, the proteomic composition of EVs affects their functionality, including alterations in proteins related to angiogenesis, coagulation, and inflammation as well as other intercellular processes essential for GBM progression [2627]. For example, GBM cell-derived EVs can stimulate cancer [28] and endothelial cell proliferation [29] during angiogenesis. Moreover, upregulation of invasive proteins in GBM cell-derived EVs, such as proteases (e.g., cathepsin D) and adhesion proteins (e.g., integrins) for extracellular matrix (ECM) interactions, favors GBM invasion [30]. As EV proteome which harbors signatures about the status of parental cells is affected by GBM progression [29313233], its proteomic nature provides causative clues regarding GBM pathogenesis. Particularly, cancer-specific EVs represent detectable materials offering another window for diagnostic opportunities related to liquid biopsy obtained from cerebrospinal fluid (CSF) or circulating blood. Thus, accurate EV proteome identification could provide a detailed biological understanding of abnormal intercellular communication in a complex GBM microenvironments and reliable biomarkers to monitor GBM progression.
Because EVs have unique size, density, protein markers, and physical properties of proteolipids, numerous biochemical methods have been used for EV isolation, including ultracentrifugation, density gradient ultracentrifugation, size exclusion chromatography, immuno-affinity purification, asymmetric flow-field-flow fractionation, and acoustics (Fig. 1) [343536]. Purity is the most important factor when selecting the isolation method. Webber and Clayton [37] suggested an excellent and straightforward method to estimate the purity of EVs based on the ratio of particle number to protein concentration. Particle number concentration of EVs is quantified by nanoparticle tracking analysis and tunable resistive pulse sensing [38]. Additionally, the recent nano-flow cytometry technology provides quantitative information regarding the number of EVs [39]. Amount of proteins in EVs is easily quantified by canonical protein quantitation methods, such as the bicinchoninic acid assay. Based on Clayton’s research, ratios over 3×1010 particles/µg are defined as high vesicular purity, ratios from 2×109 to 2×1010 particles/µg indicate low purity, and ratios below 1.5×109 particles/µg are impure [39]. High-purity EVs can be obtained from in vitro culture medium by applying sucrose cushion ultracentrifugation, while isolated low purity EVs are obtained by simple ultracentrifugation [39]. Alternatively, high-purity EVs were acquired by iodixanol density gradient ultracentrifugation of glioma cells in which 3.53×1010 particles/µg of EVs were measured [40]. Jeppesen et al. [12] employed a high-resolution iodixanol density gradient ultracentrifuge with a combination of ultracentrifugation and density gradient ultracentrifugation for isolation of high-purity exosome. Based on current knowledge, density gradient ultracentrifugation is considered the gold standard method to isolate high-purity EVs [41].
Although Clayton’s criteria for EV purity applies to in vitro culture media, in vivo biological fluids are not well validated by it. The same ultracentrifuge coupled with a washing step in urine and serum was applied but the purities of urine EVs and serum EVs were approximately 1×109 particles/µg and 7×109 particles/µg, respectively, both of which were lower than that of in vitro EVs from culture medium which was about 2×1010 particles/µg [40]. Contamination levels of isolated EVs is affected by protein concentration in the conditioned medium or biological fluid. Density gradient ultracentrifugation could be applied to isolate EVs with high purity in serum-free media and EV-depleted culture media, which are widely used for EV isolation in cell lines [42]. However, EVs in ascites of patients with colorectal cancer require repeated washing steps combined with sucrose cushion ultracentrifugation and density gradient ultracentrifugation for isolation of high-purity EVs [43].
Because protein concentration is very high in in vivo biological fluids, such as blood, these proteins cover the surface of EVs, generating a protein corona [44]. Abundant blood and ECM proteins, including apoproteins, immunoglobulins, and fibronectin, are attached to the EV surface, forming the corona [44]. Importantly, these proteins are not simple contaminants but are closely associated with EV functionality and uptake [4546]. For example, fibronectin on EVs favors their uptake by cancer cells via macropinocytosis by interacting with cellular heparin sulfate proteoglycan [46]. Thus, high degree of attachment of corona proteins on in vivo EVs makes it difficult to isolate EVs using applicable in vitro EV isolation methods. Hoshino et al. [47] applied ultracentrifugation with repeated washing steps to isolate EVs from different cell lines, tissues, blood, and other biological fluids. While they did not compare the purity level among the isolated EVs using the same procedure, EVs derived from plasma and serum represented lower number of identifications with a lower frequency of canonical EV proteins, including CD63, CD81, and tumor susceptibility gene 101 (TSG101), than EVs derived from in vitro cell lines [47], implying that contamination levels are different between EVs isolated from culture media and blood. Because mass spectrometry-based proteomics mainly identifies relatively abundant proteins [48], a higher abundance of residual EV corona proteins or other co-purified proteins in in vivo biological fluids makes it difficult to identify integrated proteins in EVs, classically considered as canonical EV marker proteins such as tetraspanins. Thus, a high-purity EV isolation method can be effectively or differently applied in biological fluids based on the relative amount of non-EV components for the generation of a reliable EV proteome.
As previously mentioned, oncogenic pathways in GBM affect the molecular content of EVs. Thus, mass spectrometry-based proteomic analyses of EVs have successfully identified alterations in proteins related to GBM pathogenesis (Table 1). Choi et al. [40] compared EVs derived from mutant EGFRvIII-expressing U373vIII and their parental indolent glioma cell line U373. EGFRvIII induced malignant transformation of U373 cells with increased invadopodia on their cell surface and tumor-forming ability in severe combined immunodeficiency mouse model [40]. This cellular oncogenic transformation was reflected in their EVs with dramatic alteration in proteomic composition, increasing pro-invasive proteins, such as CD44, basigin (BSG), CD151, and integrins [40]. Particularly, CD44 and BSG, known as EMPIRIN, co-exist in a single U373vIII EV, as their cellular expression is also co-localized in cell surface invadopodia, implying that EVs represent their parental cellular status. Thus, proteomics of EVs provides information about oncogenic changes for applicable biomarkers in GBM. Mallawaaratchy et al. [30] extensively analyzed EV proteomes from six GBM cell lines: A172, LN229, U87MG, U251, T98G, and CCF-STTG1. They also found that invasion-related EV proteins such as ANXA1, cathepsin D (CTSD), and integrin beta-1 (ITGB1) were significantly upregulated in GBM [30]. These upregulated proteins in GBM EVs were similarly observed in EVs derived from cavitron ultrasonic surgical aspirator fluid in high-grade GBM [30]. A similar study also revealed EV proteomes from five GBM cell lines, including A172, Glia-Tr, Glia-L, Glia-R, and Glia-Sh [49]. These EV proteomes also commonly identified pro-invasive proteins, such as annexin A2 (ANXA2), CD44, and tenascin-C.
GBM-derived EVs are highly equipped with invasive membrane proteins, such as CD44, melanoma cell adhesion molecule (MCAM), thrombospondin 1 (THBS1), and integrin α6β4, which play a role in tumor dissemination and interaction with ECM (Fig. 2) [40]. CD44 interacts with hyaluronic acid, which is highly overexpressed in the brain of malignant GBM patients [50]. Hyaluronic acid-rich microenvironment retains water-generating hydrogels, which favors GBM invasion [51]. Additionally, MCAM interacts with laminin, a ligand for integrin α6β4 which is implicated in organ-specific targeting of circulating EVs [5253] and a major component of basement membranes in the blood vessels of the brain tumor microenvironment [54]. Thus, GBM EVs interact with ECM in the brain tumor microenvironment and modulate GBM invasiveness. Moreover, GBM EVs have proteolytic activity similar to proteases, including disintegrin and metalloproteinase domain-containing protein 9 (ADAM9), ADAM10, cathepsin Z (CTSZ), and matrix metalloproteinase-14 (MMP14), which are involved in ECM component (laminins and collagens) degradation [40]. The cooperative action of membrane proteins and proteases in EVs could enhance their ability to bind to specific ECM and degrade selective ECM proteins [55], thereby contributing to signaling and stromal responses and facilitating GBM cell invasion [56]. Excluding cancer invasion functionality, EVs play multiple roles in the biology of GBM and tumor microenvironment [5758]; however, these aspects are not well addressed in this review.
Cancer-specific EV components offer unique diagnostic opportunities in liquid biopsy through recovery of GBM EVs from CSF or blood [59]. GBM EVs generated in the brain tend to circulate into the bloodstream across the blood-brain barrier [60]. Indira Chandran et al. [61] identified syndecan-1 (SDC1) in EVs using a proteomic approach in patient plasma as discriminative biomarker between high-grade (grade IV) and low-grade (grade II) glioma, implying that proteins in EVs are reliable biomarkers for minimally invasive diagnosis of GBM in blood. Hallar et al. [62] identified 298 differentially regulated EV proteins from neurosurgical aspirates of high-grade GBM patients. Among them, molecular chaperone T-complex proteins, including CCT2, CCT3, CCT5, CCT6A, CCT7, and TCP1, are expressed at higher levels in EVs derived from GBM patients [62]. Particularly, the higher CCT6A expression is significantly related to lower survival rate [62].
Recent single-EV analyses have revealed the heterogeneity of EVs with different molecular compositions and sizes, although they are released from the same parental cells. This diversity makes it difficult to decode the complexity of GBM EVs by harnessing their various functional and diagnostic properties. To address this complexity, different technologies for single-EV analysis have been developed, including super-resolution microscopy [636465], imaging flow cytometry [66], high-resolution flow cytometry [4064676869], interferometric imaging [70], and single EV capture platform on the chip [71]. In glioma cell line U373, nano-flow cytometry revealed that released EVs are not homogeneous with a partial positivity for CD9 (about 35%) and CD81 (about 14%), which are canonical EV marker proteins [40]. Additionally, these markers were only partially co-localized in single EVs; about 65% of U373 EVs were both CD9 and CD81 negative and 7% were double positive, representing a high degree of EV combination with different surface protein cargo [40]. EV heterogeneity is affected by oncogenic transformation by EGFRvIII, a frequent mutation related to classical glioblastoma [14] with a high degree of aggressiveness, as mentioned above [27]. U373vIII cells and U373 cells with EGFRvIII overexpression generate a different repertoire of EVs with increased mesenchymal markers, including CD44 and CD151, but decreased tertraspanin CD82 [40], which is involved in tumor metastasis suppression [72].
Another decent technology, imaging flow cytometry, detects a single EV by direct imaging with an optical lens and provides high-throughput data of EV morphology and molecular diversity by fluorescent antibody labeling [7374]. Using this approach, distinctive CD9/GFAP/SVN triple-positive EVs were observed in the plasma of patients with glioma [74]. A single EV capture platform on the chip is another effective technology for diagnosing GBM by high-sensitivity detection of single EVs [7175]. Lee et al. [71] applied this chip technology for the molecular profiling of glioma-derived EV subpopulations using multiple fluorescent antibodies for selected proteins related to GBM progression. Additionally, EV heterogeneity in mouse glioblastoma was revealed by a single EV analysis detecting higher expression of non-POU domain-containing octamer-binding protein (NonO) in larger-sized EV (>0.8 µm), whereas similar gap junction alpha-1 protein (GJA1) expression was observed in both larger and smaller EVs [76]. Advances in recent single EV detection techniques have shed light on the obscured complexity of GBM EV landscapes [39]. Particularly, specific subpopulations of EVs, not all EVs, play a role in cancer progression. Thus, interpreting a combinatorial single EV distribution could provide valuable information to understand EV-mediated GBM pathogenesis.
EVs have emerged as key regulators the in tumor microenvironment. In GBM, the production and molecular composition of EVs are affected by oncogenic transformation/mutation, acquiring new functionalities associated with cancer progression, such as invasion and angiogenesis. These observations are of interest for the investigation of GBM EV proteomes. Mass spectrometry-based proteomics has discovered high-throughput information regarding EV proteins and provided thoughtful insights into the whole landscape of the vesicular proteome, in which enrichment of invasion-related proteins is particularly connected to GBM-connected perturbations. This information provides clues about the pathogenic role of GBM EVs, as well as applicable biomarkers for the diagnostic approach in liquid biopsy. Particularly, confident EV proteomes have been acquired through high-purity EV isolation using density gradient ultracentrifugation, which is considered the gold standard method in EV proteomics. However, biological activity and diagnostic information may not be uniform across all EVs in GBM, and only a selected subpopulation of EVs can be associated with GBM progression. Thus, traditional bulk analysis of EV cargo has intrinsic limitations. Recent single EV analyses, such as nano-flow cytometry, could overcome this challenge, by enabling the detection of complex landscape and dynamic transition of EV populations depending on GBM progression. Therefore, the combination of novel vesicular protein identification by proteomics and precise mapping of EV populations by single EV analysis may pave the way for understanding GBM pathogenesis and innovative diagnostic approaches to GBM progression.
Availability of Data and Material
All data generated or analyzed during the study are included in this published article.
References
1. Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014; 30:255–289. PMID: 25288114.

2. Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013; 13:1554–1571. PMID: 23401200.

3. Choi D, Spinelli C, Montermini L, Rak J. Oncogenic regulation of extracellular vesicle proteome and heterogeneity. Proteomics. 2019; 19:e1800169. PMID: 30561828.

4. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015; 25:364–372. PMID: 25683921.

5. Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983; 97:329–339. PMID: 6309857.

6. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983; 33:967–978. PMID: 6307529.

7. Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013; 126(Pt 24):5553–5565. PMID: 24105262.

8. Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008; 319:1244–1247. PMID: 18309083.

9. Bebelman MP, Bun P, Huveneers S, van Niel G, Pegtel DM, Verweij FJ. Real-time imaging of multivesicular body-plasma membrane fusion to quantify exosome release from single cells. Nat Protoc. 2020; 15:102–121. PMID: 31836866.

10. Burnier L, Fontana P, Kwak BR, Angelillo-Scherrer A. Cell-derived microparticles in haemostasis and vascular medicine. Thromb Haemost. 2009; 101:439–451. PMID: 19277403.

11. Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, et al. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr Biol. 2009; 19:1875–1885. PMID: 19896381.

12. Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment of exosome composition. Cell. 2019; 177:428–445.e18. PMID: 30951670.

13. Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014; 3:26913. PMID: 25536934.

14. Wen PY, Weller M, Lee EQ, Alexander BM, Barnholtz-Sloan JS, Barthel FP, et al. Glioblastoma in adults: a Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 2020; 22:1073–1113. PMID: 32328653.

15. Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat Rev Clin Oncol. 2017; 14:434–452. PMID: 28031556.

16. Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007; 21:2683–2710. PMID: 17974913.

17. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005; 352:987–996. PMID: 15758009.

18. Smith JS, Wang XY, Qian J, Hosek SM, Scheithauer BW, Jenkins RB, et al. Amplification of the platelet-derived growth factor receptor-A (PDGFRA) gene occurs in oligodendrogliomas with grade IV anaplastic features. J Neuropathol Exp Neurol. 2000; 59:495–503. PMID: 10850862.

19. Xu G, Li JY. CDK4, CDK6, cyclin D1, p16(INK4a) and EGFR expression in glioblastoma with a primitive neuronal component. J Neurooncol. 2018; 136:445–452. PMID: 29150788.

20. Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019; 178:835–849.e21. PMID: 31327527.

21. Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004; 64:6892–6899. PMID: 15466178.
22. Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008; 321:1807–1812. PMID: 18772396.

23. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008; 455:1061–1068. PMID: 18772890.
24. Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013; 45:1141–1149. PMID: 23917401.

25. Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013; 155:462–477. PMID: 24120142.

26. Magnus N, Garnier D, Rak J. Oncogenic epidermal growth factor receptor up-regulates multiple elements of the tissue factor signaling pathway in human glioma cells. Blood. 2010; 116:815–818. PMID: 20462964.

27. Magnus N, Garnier D, Meehan B, McGraw S, Lee TH, Caron M, et al. Tissue factor expression provokes escape from tumor dormancy and leads to genomic alterations. Proc Natl Acad Sci U S A. 2014; 111:3544–3549. PMID: 24520174.

28. Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008; 10:1470–1476. PMID: 19011622.

29. Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringnér M, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci U S A. 2013; 110:7312–7317. PMID: 23589885.

30. Mallawaaratchy DM, Hallal S, Russell B, Ly L, Ebrahimkhani S, Wei H, et al. Comprehensive proteome profiling of glioblastoma-derived extracellular vesicles identifies markers for more aggressive disease. J Neurooncol. 2017; 131:233–244. PMID: 27770278.

31. Fujii T, Sakata A, Nishimura S, Eto K, Nagata S. TMEM16F is required for phosphatidylserine exposure and microparticle release in activated mouse platelets. Proc Natl Acad Sci U S A. 2015; 112:12800–12805. PMID: 26417084.

32. Choi DS, Choi DY, Hong BS, Jang SC, Kim DK, Lee J, et al. Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. J Extracell Vesicles. 2012; 1:18704.

33. Lee HM, Choi EJ, Kim JH, Kim TD, Kim YK, Kang C, et al. A membranous form of ICAM-1 on exosomes efficiently blocks leukocyte adhesion to activated endothelial cells. Biochem Biophys Res Commun. 2010; 397:251–256. PMID: 20529672.

34. Choi DS, Kim DK, Kim YK, Gho YS. Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom Rev. 2015; 34:474–490. PMID: 24421117.

35. Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH, Chen C, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc Natl Acad Sci U S A. 2017; 114:10584–10589. PMID: 28923936.

36. Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018; 20:332–343. PMID: 29459780.

38. Koritzinsky EH, Street JM, Star RA, Yuen PS. Quantification of exosomes. J Cell Physiol. 2017; 232:1587–1590. PMID: 27018079.

39. Choi D, Montermini L, Jeong H, Sharma S, Meehan B, Rak J. Mapping subpopulations of cancer cell-derived extracellular vesicles and particles by nano-flow cytometry. ACS Nano. 2019; 13:10499–10511. PMID: 31469961.

40. Choi D, Montermini L, Kim DK, Meehan B, Roth FP, Rak J. The impact of oncogenic EGFRvIII on the proteome of extracellular vesicles released from glioblastoma cells. Mol Cell Proteomics. 2018; 17:1948–1964. PMID: 30006486.

41. Chen J, Li P, Zhang T, Xu Z, Huang X, Wang R, et al. Review on strategies and technologies for exosome isolation and purification. Front Bioeng Biotechnol. 2021; 9:811971. PMID: 35071216.

42. Choi D, Rak J, Gho YS. Isolation of extracellular vesicles for proteomic profiling. Methods Mol Biol. 2021; 2261:193–206. PMID: 33420990.

43. Choi DS, Park JO, Jang SC, Yoon YJ, Jung JW, Choi DY, et al. Proteomic analysis of microvesicles derived from human colorectal cancer ascites. Proteomics. 2011; 11:2745–2751. PMID: 21630462.

44. Tóth EÁ, Turiák L, Visnovitz T, Cserép C, Mázló A, Sódar BW, et al. Formation of a protein corona on the surface of extracellular vesicles in blood plasma. J Extracell Vesicles. 2021; 10:e12140. PMID: 34520123.

45. Wolf M, Poupardin RW, Ebner-Peking P, Andrade AC, Blöchl C, Obermayer A, et al. A functional corona around extracellular vesicles enhances angiogenesis, skin regeneration and immunomodulation. J Extracell Vesicles. 2022; 11:e12207. PMID: 35398993.

46. Choi D, Montermini L, Meehan B, Lazaris A, Metrakos P, Rak J. Oncogenic RAS drives the CRAF-dependent extracellular vesicle uptake mechanism coupled with metastasis. J Extracell Vesicles. 2021; 10:e12091. PMID: 34136107.

47. Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020; 182:1044–1061.e18. PMID: 32795414.
48. Zubarev RA. The challenge of the proteome dynamic range and its implications for in-depth proteomics. Proteomics. 2013; 13:723–726. PMID: 23307342.

49. Naryzhny S, Volnitskiy A, Kopylov A, Zorina E, Kamyshinsky R, Bairamukov V, et al. Proteome of glioblastoma-derived exosomes as a source of biomarkers. Biomedicines. 2020; 8:216.

50. Cha J, Kang SG, Kim P. Strategies of mesenchymal invasion of patient-derived brain tumors: microenvironmental adaptation. Sci Rep. 2016; 6:24912. PMID: 27108713.

51. Koochekpour S, Pilkington GJ, Merzak A. Hyaluronic acid/CD44H interaction induces cell detachment and stimulates migration and invasion of human glioma cells in vitro. Int J Cancer. 1995; 63:450–454. PMID: 7591247.

52. Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015; 527:329–335. PMID: 26524530.

53. Mu W, Rana S, Zöller M. Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia. 2013; 15:875–887. PMID: 23908589.

54. Ljubimova JY, Fujita M, Khazenzon NM, Ljubimov AV, Black KL. Changes in laminin isoforms associated with brain tumor invasion and angiogenesis. Front Biosci. 2006; 11:81–88. PMID: 16146715.

55. McCready J, Sims JD, Chan D, Jay DG. Secretion of extracellular hsp90alpha via exosomes increases cancer cell motility: a role for plasminogen activation. BMC Cancer. 2010; 10:294. PMID: 20553606.

56. Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia. 2011; 59:1169–1180. PMID: 21446047.

57. Broekman ML, Maas SLN, Abels ER, Mempel TR, Krichevsky AM, Breakefield XO. Multidimensional communication in the microenvirons of glioblastoma. Nat Rev Neurol. 2018; 14:482–495. PMID: 29985475.

58. Quail DF, Joyce JA. The microenvironmental landscape of brain tumors. Cancer Cell. 2017; 31:326–341. PMID: 28292436.

59. Zachariah MA, Oliveira-Costa JP, Carter BS, Stott SL, Nahed BV. Blood-based biomarkers for the diagnosis and monitoring of gliomas. Neuro Oncol. 2018; 20:1155–1161. PMID: 29746665.

60. Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA, et al. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009; 23:1541–1557. PMID: 19109410.

61. Indira Chandran V, Welinder C, Månsson AS, Offer S, Freyhult E, Pernemalm M, et al. Ultrasensitive immunoprofiling of plasma extracellular vesicles identifies syndecan-1 as a potential tool for minimally invasive diagnosis of glioma. Clin Cancer Res. 2019; 25:3115–3127. PMID: 30679164.

62. Hallal S, Russell BP, Wei H, Lee MYT, Toon CW, Sy J, et al. Extracellular vesicles from neurosurgical aspirates identifies chaperonin containing TCP1 subunit 6A as a potential glioblastoma biomarker with prognostic significance. Proteomics. 2019; 19:e1800157. PMID: 30451371.

63. Chen C, Zong S, Wang Z, Lu J, Zhu D, Zhang Y, et al. Imaging and intracellular tracking of cancer-derived exosomes using single-molecule localization-based super-resolution microscope. ACS Appl Mater Interfaces. 2016; 8:25825–25833. PMID: 27617891.

64. Higginbotham JN, Zhang Q, Jeppesen DK, Scott AM, Manning HC, Ochieng J, et al. Identification and characterization of EGF receptor in individual exosomes by fluorescence-activated vesicle sorting. J Extracell Vesicles. 2016; 5:29254. PMID: 27345057.

65. Lennon KM, Wakefield DL, Maddox AL, Brehove MS, Willner AN, Garcia-Mansfield K, et al. Single molecule characterization of individual extracellular vesicles from pancreatic cancer. J Extracell Vesicles. 2019; 8:1685634. PMID: 31741725.

66. Mastoridis S, Bertolino GM, Whitehouse G, Dazzi F, Sanchez-Fueyo A, Martinez-Llordella M. Multiparametric analysis of circulating exosomes and other small extracellular vesicles by advanced imaging flow cytometry. Front Immunol. 2018; 9:1583. PMID: 30034401.

67. Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D, et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J Extracell Vesicles. 2015; 4:25530. PMID: 25833224.

68. Morales-Kastresana A, Telford B, Musich TA, McKinnon K, Clayborne C, Braig Z, et al. Labeling extracellular vesicles for nanoscale flow cytometry. Sci Rep. 2017; 7:1878. PMID: 28500324.

69. Danielson KM, Estanislau J, Tigges J, Toxavidis V, Camacho V, Felton EJ, et al. Diurnal variations of circulating extracellular vesicles measured by nano flow cytometry. PLoS One. 2016; 11:e0144678. PMID: 26745887.

70. Yang Y, Shen G, Wang H, Li H, Zhang T, Tao N, et al. Interferometric plasmonic imaging and detection of single exosomes. Proc Natl Acad Sci U S A. 2018; 115:10275–10280. PMID: 30249664.

71. Lee K, Fraser K, Ghaddar B, Yang K, Kim E, Balaj L, et al. Multiplexed profiling of single extracellular vesicles. ACS Nano. 2018; 12:494–503. PMID: 29286635.

72. Liu WM, Zhang XA. KAI1/CD82, a tumor metastasis suppressor. Cancer Lett. 2006; 240:183–194. PMID: 16260083.

73. Ricklefs FL, Maire CL, Reimer R, Dührsen L, Kolbe K, Holz M, et al. Imaging flow cytometry facilitates multiparametric characterization of extracellular vesicles in malignant brain tumours. J Extracell Vesicles. 2019; 8:1588555. PMID: 30949309.

74. Galbo PM Jr, Ciesielski MJ, Figel S, Maguire O, Qiu J, Wiltsie L, et al. Circulating CD9+/GFAP+/survivin+ exosomes in malignant glioma patients following survivin vaccination. Oncotarget. 2017; 8:114722–114735. PMID: 29383115.

75. Fraser K, Jo A, Giedt J, Vinegoni C, Yang KS, Peruzzi P, et al. Characterization of single microvesicles in plasma from glioblastoma patients. Neuro Oncol. 2019; 21:606–615. PMID: 30561734.

76. Gyuris A, Navarrete-Perea J, Jo A, Cristea S, Zhou S, Fraser K, et al. Physical and molecular landscapes of mouse glioma extracellular vesicles define heterogeneity. Cell Rep. 2019; 27:3972–3987.e6. PMID: 31242427.

Fig. 1
Overview of methodological approaches to isolate EV for proteomics. Density gradient ultracentrifuge is considered the gold-standard method in EV proteomics. The purity of EVs could be estimated by particle number per protein amount as suggested by Webber and Clayton [37]. EV, extracellular vesicle; CSF, cerebrospinal fluid; NTA, nanoparticle tracking analysis; TRPS, tunable resistive pulse sensing.
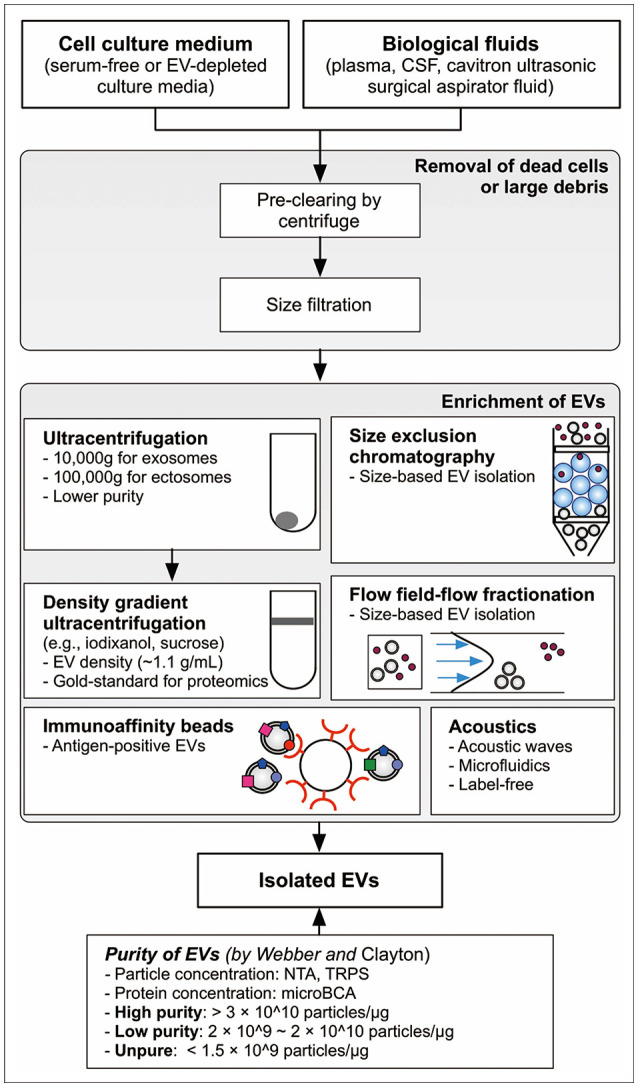
Fig. 2
EV subtypes and heterogeneity. Glioblastoma cell releases both ectosomes and exosomes. Oncogenic transformation during GBM progression alters the single EV distribution with different proteomic compositions related to cancer invasion. Moreover, because released EVs from glioblastoma cells represent the status of parental cells, decoding of EV heterogeneity and complexity by a single EV analysis could be applied in the diagnosis against GBM. EV, extracellular vesicle; GBM, glioblastoma multiforme; MVB, multivesicular body; BSG, basigin; ECM, extracellular matrix.
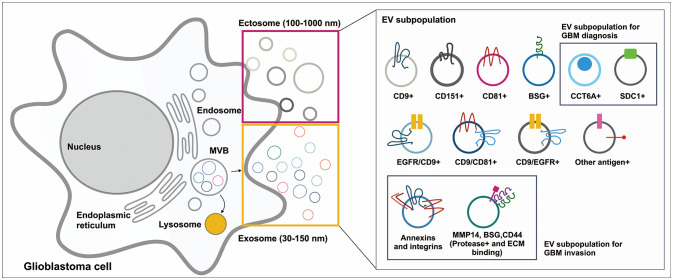
Table 1
Proteomics of EVs in GBM
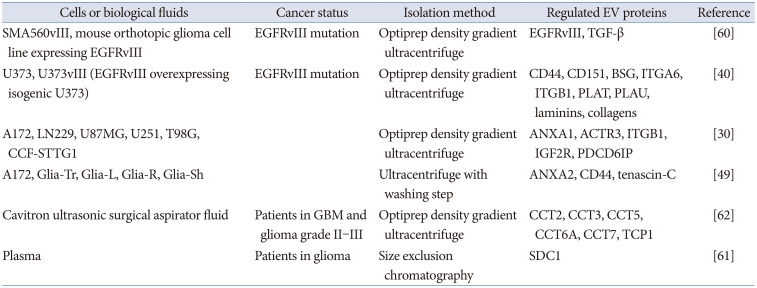
| Cells or biological fluids | Cancer status | Isolation method | Regulated EV proteins | Reference |
|---|---|---|---|---|
| SMA560vIII, mouse orthotopic glioma cell line expressing EGFRvIII | EGFRvIII mutation | Optiprep density gradient ultracentrifuge | EGFRvIII, TGF-β | [60] |
| U373, U373vIII (EGFRvIII overexpressing isogenic U373) | EGFRvIII mutation | Optiprep density gradient ultracentrifuge | CD44, CD151, BSG, ITGA6, ITGB1, PLAT, PLAU, laminins, collagens | [40] |
| A172, LN229, U87MG, U251, T98G, CCF-STTG1 | Optiprep density gradient ultracentrifuge | ANXA1, ACTR3, ITGB1, IGF2R, PDCD6IP | [30] | |
| A172, Glia-Tr, Glia-L, Glia-R, Glia-Sh | Ultracentrifuge with washing step | ANXA2, CD44, tenascin-C | [49] | |
| Cavitron ultrasonic surgical aspirator fluid | Patients in GBM and glioma grade II–III | Optiprep density gradient ultracentrifuge | CCT2, CCT3, CCT5, CCT6A, CCT7, TCP1 | [62] |
| Plasma | Patients in glioma | Size exclusion chromatography | SDC1 | [61] |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download