Abstract
Endothelial cells (EC) are the anatomical boundaries between the intravascular and extravascular space. Damage to ECs is catastrophic and induces endothelial cell dysfunction. The pathogenesis is multifactorial and involves dysregulation in the signaling pathways, membrane lipids ratio disturbance, cell-cell adhesion disturbance, unfolded protein response, lysosomal and mitochondrial stress, autophagy dysregulation, and oxidative stress. Autophagy is a lysosomal-dependent turnover of intracellular components. Autophagy was recognized early in the pathogenesis of endothelial dysfunction. Autophagy is a remarkable patho (physiological) process in the cell homeostasis regulation including EC. Regulation of autophagy rate is disease-dependent and impaired with aging. Up-regulation of autophagy induces endothelial cell regeneration/differentiation and improves the function of impaired ones. The paper scrutinizes the molecular mechanisms and triggers of EC dysregulation and current perspectives for future therapeutic strategies by autophagy targeting.
Tremendous advances in vascular biology have been achieved in recent years. Physiologically, the endothelium comprises highly specialized cells that line the blood and lymphatic vessels, weigh 1 kg and build approximately 400 m2 in an adult person that maintains approximately 60,000 miles of blood vessels [1]. Endothelial cells (EC) performs a wide range of crucial metabolic homeostatic activities to maintain a balance between aggression and protection factors by responding to extrinsic and intrinsic signals and releasing specific active molecules, as well as remodeling signaling pathways [1-3]. Endothelial cell homeostasis is crucial for maintaining the functionality of the whole organism. Classically, EC regulate vascular permeability, angiogenesis, immune system, pro- and anti-coagulant system, promotion of vasodilation, antioxidant effects, anti-inflammatory effects, inhibition of leukocyte adhesion and migration, inhibition of smooth muscle cell proliferation and migration, inhibition of platelet aggregation and adhesion, profibrinolytic effects, and regeneration of tissues [4].
Autophagy belongs to the four main types of cell death; apoptosis, autophagy, necrosis, and entosis [5]. However, stressed EC experience one of the following types of cell death, apoptosis, autophagy, or pyroptosis [6, 7]. According to the pathophysiological changes in the molecular signaling pathway and the triggering molecules, and severity of oxidative stress as well as the likelihood of endothelial cell to survive, the EC decide the most appropriate pathway of cell death. EC apoptosis is upregulated during oxidative stress, nutrient starvation, tumor necrosis factor-α (TNF-α), high glucose, lipopolysaccharide, homocysteine [8]. However, the full mechanism of regulation of endothelial cell death and the decision on the death pathway is not fully studied.
Vascular EC impairment involved in the pathogenesis of various diseases, including sepsis, diabetes, cerebral ischemia, cardiac ischemia/reperfusion injury, atherosclerosis, and chronic kidney disease [9-11]. Therefore, the analysis of the function of the endothelial cell is crucial and involved in the treatment strategy of such common diseases.
Several kinds of basal autophagy present in norm including macroautopahgy, microautopahgy, and chaperon mediated autophagy. Macroautophagy is the most explored type, which involves the turnover of intracellular components such as proteins, lipids, and organelles, as well as micronucleophagy. Selective autophagy targeting induces safer and more effective therapeutic results.
Endothelial cell damage can be induced by uncontrolled external mechanical shear stress and contractile forces such as mechanical stretching and vascular wall stress (biologically uncontrolled accidental cell death) or by regulated cell death [6]. Autophagy is a classical variant of regulated cell death. Usually, endothelial damage occurs due to mechanical shear stress and damage to the cell wall membrane. Whereas, regulated cell death (autophagy) is the primary target of future therapeutic strategy. Typically, early stages of regulated cell death are functional endothelial cell dysfunction. The boundaries between the reversible and irreversible change tends to be crucial during development of new therapeutic strategy for endothelial cell dysfunction.
Induction of endothelial cell damage is multifactorial and involved several pathways, including activation of autophagy stimulatory signaling pathways (miRNA-126, Apelin), inhibition of inhibitory signaling pathways (Notch1), stimulation of autophagy inducing receptors, inhibition of inhibitory receptors (caspase inhibitor Z-VAD-FMK, autophagy inhibitor 3-MA) and activation of autophagy inducing molecules and inhibiting autophagy inhibitory molecules [12-15].
Each of these has its role in the pathogenesis of endothelial cell autophagy dysregulation that subsequently results in endothelial cell dysfunction. Usually, a combination of these disorders coexists.
Cytokines and chemokines are the leading cause of endothelial cell dysfunction. Interleukin-6 (IL-6) is one of the first cytokines studied that induces endothelial cell dysfunction and the release of the endothelial plasminogen activator inhibitor-1 [16]. Classically, IL-6 levels are elevated during sepsis, bacterial infection, and acute respiratory distress syndrome. Hyperproduction of pro-inflammatory cytokines such as TNF-α, IL-8, and monocyte chemotactic protein-1 leads to cytokine release syndrome [16]. Cytokine release syndrome is a major stress to EC, prolonged exposure of EC to pro-inflammatory mediators.
Generally, degradation of intracellular debridement’s can be achieved through microautophagy, macroautophagy, and chaperone-mediated autophagy depending on the debridement marker flag [17]. Endothelial cell autophagy mechanism is identical to that of myocardiocytes. However, the molecular triggers and inhibitors are different. Classically, autophagy is regulated by several autophagy-related proteins such as systems that produce modified complexes Atg8-PE and Atg5-Atg12-Atg16 [18]. More than 35 autophagy-related protein (Atg) genes have been identified, 17 of which are indispensable.
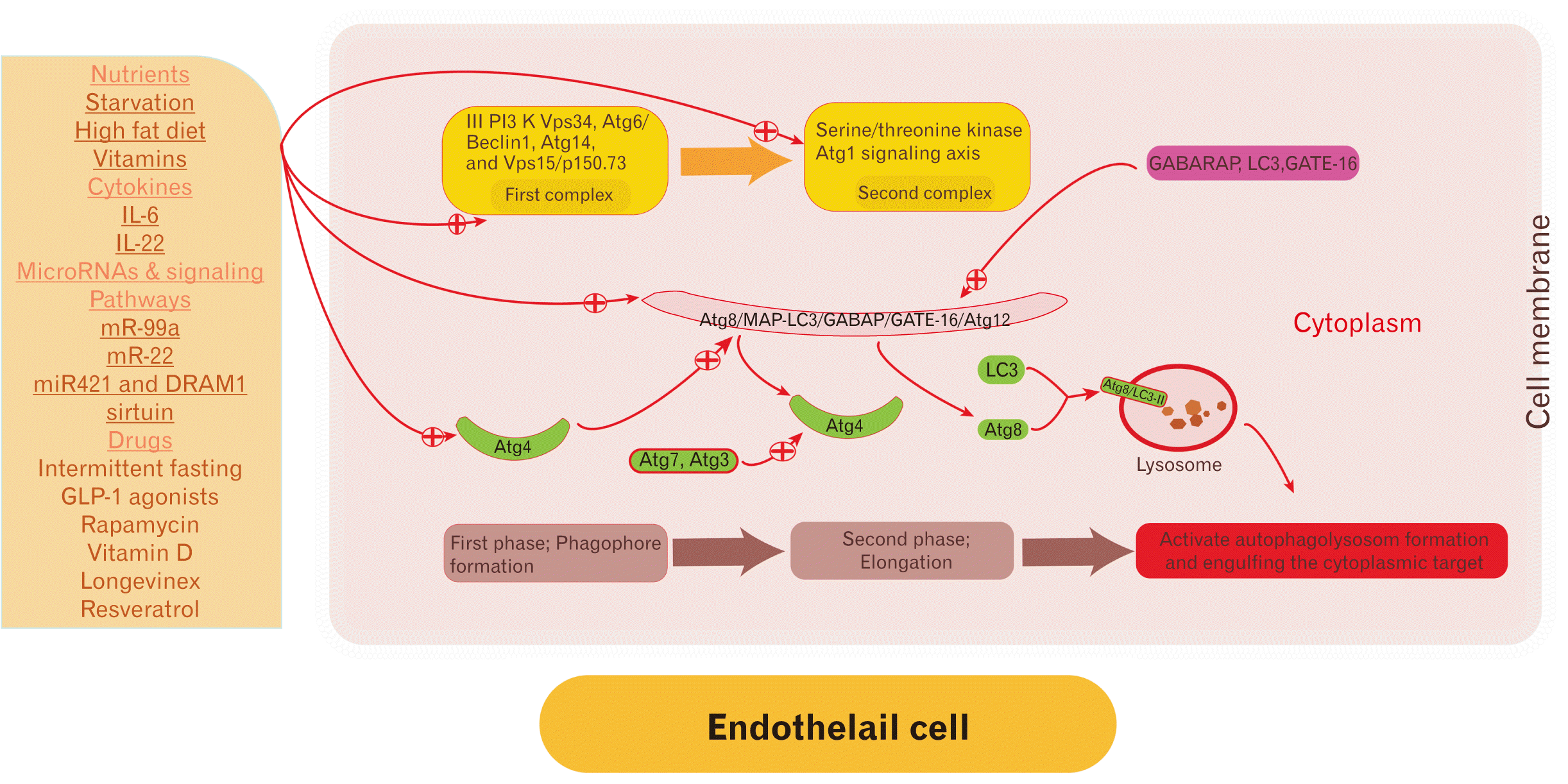
Mechanically, endothelial cell autophagy involves -Atg14L-Vps15-Vps34 proteins. Phagophore is the first phase of autophagosome formation. Two primary complexes signaling pathways involved in autophagosome formation [19, 20]. Autophagosome initiation induced by unc-51 like autophagy activating kinase 1 (Atg1) or by phosphatidylinositol 3-kinase (PI3K) Vps34 with Atg6 [21]. Autophagosome distention is regulated by transcription and post-transcription modulations, and epigenetics.
Initially, the first complex is composed of III PI3K Vps34, Atg6/Beclin1, Atg14, and Vps15/p150.73. Whereas the second complex mainly involves the serine/threonine kinase Atg1 signaling axis that its functionally depends on the presence of LC3, GATE-16 and GABARAP proteins [18].
Subsequently, autophagy elongation occurs that involves ubiquitin-like conjugation pathways; Atg8/MAP-LC3/GABAP/GATE-16 and Atg12 systems. Thereafter, the Atg8 protein undergoes cleavage for its carboxyl-terminal by the cysteine protease Atg4. Then, Atg4 undergoes a long complexed signaling cascade involved activation of Atg4 by Atg7 and Atg3 that culminate in activation of Atg8 and it is binding with the LC3–II of the lysosome and formation of Atg8/LC3–II complex. The Atg8/LC3–II complex bind with autophagosome in a covalent bound unless Atg4 cleaves it to be recycled and degraded by the phagolysosome [18]. In human, the fusion of the autophagosome with the lysosome remains not fully understood. Small GTPase Rab7 and lysosomal acidification through lysosomal H+-ATPase are required for successful fusion (Fig. 1).
Autolysosome formation regulated by genetic/epigenetic/transcriptional/posttranscriptional modifications [22].
Induction of endothelial cell autophagy occurs physiologically under hypoxic conditions. Several mechanistic pathways are potentially possible to take place, Inhibition of mTOR receptor, proapoptotic gene Bnip3, IP3 and lysosome inhibitors depending on the hypoxia severity. During anoxia, hypoxia-inducible factor 1, α subunit (HIF1A) is activated. HIF1A induces the expression of the Bnip3 gene (encode for BH3-only protein) which is required for disrupting the BCL2–BECN1 interaction by competitive binding of BCL2 (B-cell CLL/lymphoma 2) [23]. Bnip3 is an important gene for activating autophagy during hypoxic and non-hypoxic states.
The pathogenesis of endothelial dysfunction is multifactorial and involves chronic mild inflammatory reactions, direct physical stimulation and shear stress, chemical poisons and cytokine/chemokine release syndrome, concurrency of related diseases, and molecular changes on the cellular and subcellular level. The cell in rule has a precise equilibrium between the protective and aggression factors. Disruption to the natural balance and shifting the balance to the aggression side induces local endothelial dysfunction. Subsequently, over time, the prolonged predominant of the aggression factors resulted in multi-organ systemic changes in endothelial cell function. Development of an effective tool to identify the early stage of local endothelial dysfunction is crucial to prevent further progression into systemic multiorgan endothelial cell dysfunction. Seemingly, first place where endothelial cell get dysfunction and damage is in the branches and bifurcations and in regions of sharp vessel curvature where high shear stress. Early detection of level of the endothelial dysfunction markers such as E-selectin, ICAM-1, and VCAM-1, endoglin, endocan, von Willebrand factor, and soluble thrombomodulin is crucial to evaluate the risk of development of complications such as atherosclerosis [24-32]. Changes in the endothelial cell predicts the risk of endothelial cell injury. Recent study formulated an equation to early asses the risk of cardiovascular disease development depending on the flow-mediated dilation (FMD) of the arteries (FMD=–0.096BMI–0.069age–4.551WHR–0.015AST–0.242LDL+17.938) [33].
The most likely hypothesis is that in obese people with dyslipidemia, hyperglycemia, and hypertension, the EC develop functional memory and overtime reduces its response to the changes in the glucose level, blood pressure, and lipid profile. Persistent fluctuations in the metabolic syndrome component alter the functionality, biosynthesis, and response of EC to external stimuli. Prolonged changes results in mild chronic inflammation of the EC and the underlying smooth muscle cells. A vicious circle develops in the background each time the endothelial cell activity is impaired more and in the final stage there are anatomical and topographical changes in the EC. loss of endothelial cell function reduces the bioavailability of nitric oxide which subsequently leads to persistent vascular smooth muscle constriction and hypertrophy of the vascular smooth muscle cells [34]. Final outcome of this process is vascular damage and loss vascular tone regulation.
According to the topographical anatomy of EC, vascular and lymphatic endothelial cell exist. Several fundamental biological distinctions are present between lymphatic and blood EC despite the topographical similarities, more than 400 differences in the expressed genes, returning to the difference in the functions of each (Table 1) [39-46]. Lymphatic EC present semiprofessional antigen presentation to lymphatic circulatory T cytotoxic CD8 cells through an invadosome-shaped protrusion and express specific surface antigens and receptors such as podoplanin, vascular endothelial growth factor receptor-3 (VEGFR-3) or lymphatic vessel endothelial hyaluronan receptor-1 [47, 48].
Remarkable evidence concluded that sigma receptor-1 of the lymphatic EC is responsible for the regulation of the tone and barrier function as well as the permeability of the lymphatic tree through regulation of nitroxide biosynthesis signaling pathway [49].
Recent meta-analysis showed approximately, 400 differentially expressed genes, 36 core-features, and 57 core-pathways have been found to be associated with ECs senescence [50]. Moreover, the phosphoglycerate dehydrogenase and the serine biosynthesis pathway are the predominant in EC senescence [50]. Impaired autophagy is a hallmark during aging of EC. Early evaluation of the level of the autophagy genes suggests the biological age of the cells, in addition with using the epigenome as a reliable tool to identify the biological age of the cells and particularly the endothelial cell (Fig. 2).
Endothelial cell dysfunction is a global issue requires massive attention. Current therapeutic strategies aimed to reverse the pathogenetic pathway and return the physio-anatomy of the EC. However, targeting autophagy as a therapeutic strategy remains in the early stages of study.
The therapeutic strategy includes lifestyle modification, antioxidant, autophagy inducers, control component of metabolic syndrome (dyslipidemia, hypertension, hyperglycemia), epigenetic modification, genetic modifications, transcription factors, DNA methylation or demethylation, microenvironment correction, cell-cell adhesion molecules correction.
Pharmacotherapy such as Resveratrol reduces endothelial cell dysfunction by modulating Bnip3 associated mitophagy [52]. Several other medications are in preclinical and clinical trials to assess their therapeutic value in the regulation of autophagy, such as Metformin, Rapamycin, Sirtuin, Velcade, Artenimol, and Longevinex [8].
Transcription factors has been seen to induce endothelial cell regeneration in animal models particularly pulmonary vessels EC by FoxM1 transcription factor [52]. The ability of the transcription factors (Oct4 and Klf4) to transdifferentiate other non-EC such as fibroblast into EC has shown promising results [53]. Moreover, upregulation of autophagy can be achieved by modification of autophagy gene regulation through transcription factors. Hypoxia-inducible factors-α induces endothelial cell regeneration in animal models through the FoxM1 transcription factor [54]. Sox17 is another specific TF for EC proliferation shows stimulatory effect when its expression upregulated [55]. Atf3 is another key transcription factor in the regeneration strategy of damaged EC [56]. These transcription factors induce common signaling pathways between autophagy and cell cycle regulation encoding genes. Therefore, autophagy induction induces cell regeneration and improves the functionality of impaired EC. Regeneration of EC by the transcription factors culminates in one common result. FoxM1 induces regeneration through upregulation the signaling G-coupled protein receptor–dependent p110γ isoform of phosphoinositide 3-kinase (PI3K). Coexistence of FoxM1 and Pik3cg expression enhances the regeneration capacity of the EC [57]. Several other signaling pathways involved in the regeneration of EC such as Apelin signaling, Notch1 signaling, miRNA-126P signaling, VEGF signaling, and insulin-like growth factor (IGF)-1 receptor (IGF1R) [14, 58-61].
Autophagy plays a pivotal role in the pathogenesis of endothelial cell dysfunction. Where basal autophagy is required to prevent endothelial cell damage and survive the stress conditions as well as prevent endothelial cell reprogramming. However, reduced basal autophagy level is indirect cause of endothelial dysfunction and cell death. Majorly, mitophagy is impaired during endothelial cell dysfunction which is responsible for oxidative stress alleviating and recycling impaired mitochondria [62, 63].
Currently, several invasive and non-invasive methods are known to evaluate the functionality of EC in different parts of the vascular tree. Of these methods are venous occlusion plethysmography, flow-mediated dilatation, pulse wave analysis, peripheral arterial tonometry, laser Doppler flowmetry, biochemical markers and bioassays, biochemical markers and bioassays, endothelial microparticles, endothelial progenitor cells, endothelial progenitor cells, and endothelial glycocalyx [64]. Autophagy interferes with apoptosis signaling pathway regulation and several molecules can interact with autophagy and prevent apoptosis and induces necrosis of EC [65-67]. Apoptosis involved common caspase signaling pathway, blocking of caspases activity induces regulated necrosis such as in endothelial cell; pyroptosis [68].
The impairment of endothelial cell autophagy is the basis for endothelial dysfunction. Autophagy does not only self-eating the intracellular component; autophagy effectively participates in maintaining endothelial cell homeostasis. Autophagy prevents cell linage transfer, stabilizes mitochondrial function subsequent protection from oxidative stress, improves cellular antioxidant defense system, prevents endoplasmic reticulum stress, maintains intracellular cell signaling, regulation cell-cell adhesion, enhances proliferation and further angiogenesis, promotes genomic stability of mitochondria and the nucleus, improves transmembrane transportation including mitochondria permeability transition, improves endothelial cell adaptation to macro and microenvironment changes, and improves the synthetic and release function of impaired endothelial cell [68].
Notes
References
1. Bierhansl L, Conradi LC, Treps L, Dewerchin M, Carmeliet P. 2017; Central role of metabolism in endothelial cell function and vascular disease. Physiology (Bethesda). 32:126–40. DOI: 10.1152/physiol.00031.2016. PMID: 28202623. PMCID: PMC5337830.

2. Khan S, Taverna F, Rohlenova K, Treps L, Geldhof V, de Rooij L, Sokol L, Pircher A, Conradi LC, Kalucka J, Schoonjans L, Eelen G, Dewerchin M, Karakach T, Li X, Goveia J, Carmeliet P. 2019; EndoDB: a database of endothelial cell transcriptomics data. Nucleic Acids Res. 47(D1):D736–44. DOI: 10.1093/nar/gky997. PMID: 30357379. PMCID: PMC6324065.

3. Rajendran P, Rengarajan T, Thangavel J, Nishigaki Y, Sakthisekaran D, Sethi G, Nishigaki I. 2013; The vascular endothelium and human diseases. Int J Biol Sci. 9:1057–69. DOI: 10.7150/ijbs.7502. PMID: 24250251. PMCID: PMC3831119.

4. Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. 2008; The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 15:261–71. DOI: 10.1016/j.devcel.2008.07.002. PMID: 18694565. PMCID: PMC2685763.

5. Chen Y, Hua Y, Li X, Arslan IM, Zhang W, Meng G. 2020; Distinct types of cell death and the implication in diabetic cardiomyopathy. Front Pharmacol. 11:42. DOI: 10.3389/fphar.2020.00042. PMID: 32116717. PMCID: PMC7018666. PMID: bd22dd15e8ff4ec4beacaf4aa092b797.

6. Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, Annicchiarico-Petruzzelli M, Antonov AV, Arama E, Baehrecke EH, Barlev NA, Bazan NG, Bernassola F, Bertrand MJM, Bianchi K, Blagosklonny MV, Blomgren K, Borner C, Boya P, Brenner C, Campanella M, Candi E, Carmona-Gutierrez D, Cecconi F, Chan FK, Chandel NS, Cheng EH, Chipuk JE, Cidlowski JA, Ciechanover A, Cohen GM, Conrad M, Cubillos-Ruiz JR, Czabotar PE, D'Angiolella V, Dawson TM, Dawson VL, De Laurenzi V, De Maria R, Debatin KM, DeBerardinis RJ, Deshmukh M, Di Daniele N, Di Virgilio F, Dixit VM, Dixon SJ, Duckett CS, Dynlacht BD, El-Deiry WS, Elrod JW, Fimia GM, Fulda S, García-Sáez AJ, Garg AD, Garrido C, Gavathiotis E, Golstein P, Gottlieb E, Green DR, Greene LA, Gronemeyer H, Gross A, Hajnoczky G, Hardwick JM, Harris IS, Hengartner MO, Hetz C, Ichijo H, Jäättelä M, Joseph B, Jost PJ, Juin PP, Kaiser WJ, Karin M, Kaufmann T, Kepp O, Kimchi A, Kitsis RN, Klionsky DJ, Knight RA, Kumar S, Lee SW, Lemasters JJ, Levine B, Linkermann A, Lipton SA, Lockshin RA, López-Otín C, Lowe SW, Luedde T, Lugli E, MacFarlane M, Madeo F, Malewicz M, Malorni W, Manic G, Marine JC, Martin SJ, Martinou JC, Medema JP, Mehlen P, Meier P, Melino S, Miao EA, Molkentin JD, Moll UM, Muñoz-Pinedo C, Nagata S, Nuñez G, Oberst A, Oren M, Overholtzer M, Pagano M, Panaretakis T, Pasparakis M, Penninger JM, Pereira DM, Pervaiz S, Peter ME, Piacentini M, Pinton P, Prehn JHM, Puthalakath H, Rabinovich GA, Rehm M, Rizzuto R, Rodrigues CMP, Rubinsztein DC, Rudel T, Ryan KM, Sayan E, Scorrano L, Shao F, Shi Y, Silke J, Simon HU, Sistigu A, Stockwell BR, Strasser A, Szabadkai G, Tait SWG, Tang D, Tavernarakis N, Thorburn A, Tsujimoto Y, Turk B, Vanden Berghe T, Vandenabeele P, Vander Heiden MG, Villunger A, Virgin HW, Vousden KH, Vucic D, Wagner EF, Walczak H, Wallach D, Wang Y, Wells JA, Wood W, Yuan J, Zakeri Z, Zhivotovsky B, Zitvogel L, Melino G, Kroemer G. 2018; Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25:486–541. DOI: 10.1038/s41418-017-0012-4. PMID: 29362479. PMCID: PMC5864239.

7. Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, Hengartner M, Knight RA, Kumar S, Lipton SA, Malorni W, Nuñez G, Peter ME, Tschopp J, Yuan J, Piacentini M, Zhivotovsky B, Melino G. 2009; Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 16:3–11. DOI: 10.1038/cdd.2008.150. PMID: 18846107. PMCID: PMC2744427.

8. Marzoog BA, Vlasova TI. 2022; Myocardiocyte autophagy in the context of myocardiocytes regeneration: a potential novel therapeutic strategy. Egypt J Med Hum Genet. 23:41. DOI: 10.1186/s43042-022-00250-8. PMID: d1da3ab8dd21477095572d54dc7e6bc3.

9. Joffre J, Hellman J, Ince C, Ait-Oufella H. 2020; Endothelial responses in sepsis. Am J Respir Crit Care Med. 202:361–70. DOI: 10.1164/rccm.201910-1911TR. PMID: 32101446.

10. Paone S, Baxter AA, Hulett MD, Poon IKH. 2019; Endothelial cell apoptosis and the role of endothelial cell-derived extracellular vesicles in the progression of atherosclerosis. Cell Mol Life Sci. 76:1093–106. DOI: 10.1007/s00018-018-2983-9. PMID: 30569278.

11. Morris G, Puri BK, Olive L, Carvalho A, Berk M, Walder K, Gustad LT, Maes M. 2020; Endothelial dysfunction in neuroprogressive disorders-causes and suggested treatments. BMC Med. 18:305. DOI: 10.1186/s12916-020-01749-w. PMID: 33070778. PMCID: PMC7570030. PMID: 2ab20273ea0c4e84a8a10b83ba3bcfa9.

12. Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RT, Heyll K, Noels H, Hristov M, Wang S, Kiessling F, Olson EN, Weber C. 2014; MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med. 20:368–76. DOI: 10.1038/nm.3487. PMID: 24584117. PMCID: PMC4398028.

13. Jansen F, Yang X, Hoelscher M, Cattelan A, Schmitz T, Proebsting S, Wenzel D, Vosen S, Franklin BS, Fleischmann BK, Nickenig G, Werner N. 2013; Endothelial microparticle-mediated transfer of MicroRNA-126 promotes vascular endothelial cell repair via SPRED1 and is abrogated in glucose-damaged endothelial microparticles. Circulation. 128:2026–38. DOI: 10.1161/CIRCULATIONAHA.113.001720. PMID: 24014835.

14. Masoud AG, Lin J, Azad AK, Farhan MA, Fischer C, Zhu LF, Zhang H, Sis B, Kassiri Z, Moore RB, Kim D, Anderson CC, Vederas JC, Adam BA, Oudit GY, Murray AG. 2020; Apelin directs endothelial cell differentiation and vascular repair following immune-mediated injury. J Clin Invest. 130:94–107. DOI: 10.1172/JCI128469. PMID: 31738185. PMCID: PMC6934203.

15. Xia W, Yin J, Zhang S, Guo C, Li Y, Zhang Y, Zhang A, Jia Z, Chen H. 2018; Parkin modulates ERRα/eNOS signaling pathway in endothelial cells. Cell Physiol Biochem. 49:2022–34. DOI: 10.1159/000493713. PMID: 30244249.

16. Kang S, Tanaka T, Inoue H, Ono C, Hashimoto S, Kioi Y, Matsumoto H, Matsuura H, Matsubara T, Shimizu K, Ogura H, Matsuura Y, Kishimoto T. 2020; IL-6 trans-signaling induces plasminogen activator inhibitor-1 from vascular endothelial cells in cytokine release syndrome. Proc Natl Acad Sci U S A. 117:22351–6. DOI: 10.1073/pnas.2010229117. PMID: 32826331. PMCID: PMC7486751.

17. Cuervo AM, Wong E. 2014; Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 24:92–104. DOI: 10.1038/cr.2013.153. PMID: 24281265. PMCID: PMC3879702.

18. Suzuki K, Ohsumi Y. 2007; Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 581:2156–61. DOI: 10.1016/j.febslet.2007.01.096. PMID: 17382324.
19. Dikic I, Elazar Z. 2018; Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 19:349–64. DOI: 10.1038/s41580-018-0003-4. PMID: 29618831.

20. Gatica D, Chiong M, Lavandero S, Klionsky DJ. 2015; Molecular mechanisms of autophagy in the cardiovascular system. Circ Res. 116:456–67. Erratum in: Circ Res 2015;116:e56. DOI: 10.1161/CIRCRESAHA.114.303788. PMID: 25634969. PMCID: PMC4313620.

21. Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. 2010; Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 90:1383–435. DOI: 10.1152/physrev.00030.2009. PMID: 20959619.

22. Di Malta C, Cinque L, Settembre C. 2019; Transcriptional regulation of autophagy: mechanisms and diseases. Front Cell Dev Biol. 7:114. DOI: 10.3389/fcell.2019.00114. PMID: 31312633. PMCID: PMC6614182.

23. Hale AN, Ledbetter DJ, Gawriluk TR, Rucker EB 3rd. 2013; Autophagy: regulation and role in development. Autophagy. 9:951–72. DOI: 10.4161/auto.24273. PMID: 24121596. PMCID: PMC3722331.
24. Icli A, Cure E, Cure MC, Uslu AU, Balta S, Mikhailidis DP, Ozturk C, Arslan S, Sakız D, Sahin M, Kucuk A. 2016; Endocan levels and subclinical atherosclerosis in patients with systemic lupus erythematosus. Angiology. 67:749–55. DOI: 10.1177/0003319715616240. PMID: 26614790.

25. Leite AR, Borges-Canha M, Cardoso R, Neves JS, Castro-Ferreira R, Leite-Moreira A. 2020; Novel biomarkers for evaluation of endothelial dysfunction. Angiology. 71:397–410. DOI: 10.1177/0003319720903586. PMID: 32077315.

26. Musialowska D, Zbroch E, Koc-Zorawska E, Musialowski P, Malyszko J. 2018; Endocan concentration in patients with primary hypertension. Angiology. 69:483–9. DOI: 10.1177/0003319717736158. PMID: 29084442.

27. Oral E, Halici Z, Cinar I, Ozcan E, Kutlu Z. 2019; Evaluation of endothelial dysfunction in bipolar affective disorders: serum endocan and urotensin-II levels. Clin Psychopharmacol Neurosci. 17:211–21. DOI: 10.9758/cpn.2019.17.2.211. PMID: 30905121. PMCID: PMC6478082.

28. Ibrahim SMA, Sabah HMA, Eldesoky AI, Soltan MY, Elshamy HA, Abdulhady H. 2022; Role of endothelial cell specific molecule-1 (endocan) and cardiac imaging in early detection of cardiac involvement in psoriatic patients with or without arthritis. Egypt Rheumatol. 44:159–64. DOI: 10.1016/j.ejr.2021.11.002.

29. Altintas N, Mutlu LC, Akkoyun DC, Aydin M, Bilir B, Yilmaz A, Malhotra A. 2016; Effect of CPAP on new endothelial dysfunction marker, endocan, in people with obstructive sleep apnea. Angiology. 67:364–74. DOI: 10.1177/0003319715590558. PMID: 26076702. PMCID: PMC4846580.

30. Balamir I, Ates I, Topcuoglu C, Turhan T. 2018; Association of endocan, ischemia-modified albumin, and hsCRP levels with endothelial dysfunction in type 2 diabetes mellitus. Angiology. 69:609–16. DOI: 10.1177/0003319717740781. PMID: 29172652.

31. Çimen T, Efe TH, Akyel A, Sunman H, Algül E, Şahan HF, Erden G, Özdemir Ş, Alay EF, Doğan M, Yeter E. 2016; Human endothelial cell-specific molecule-1 (Endocan) and coronary artery disease and microvascular angina. Angiology. 67:846–53. DOI: 10.1177/0003319715625827. PMID: 26744512.

32. Aparci M, Isilak Z, Uz O, Yalcin M, Kucuk U. 2015; Endocan and endothelial dysfunction. Angiology. 66:488–9. DOI: 10.1177/0003319714568791. PMID: 25632053.

33. Li X, Liu H, Zhang Y, Gu Y, Sun L, Yu H, Bai W. 2022; A prediction equation to estimate vascular endothelial function in different body mass index populations. Front Cardiovasc Med. 9:766565. DOI: 10.3389/fcvm.2022.766565. PMID: 35360015. PMCID: PMC8960173. PMID: 68c07f4677044d7b9b7da88a3981e63a.

34. Mudau M, Genis A, Lochner A, Strijdom H. 2012; Endothelial dysfunction: the early predictor of atherosclerosis. Cardiovasc J Afr. 23:222–31. DOI: 10.5830/CVJA-2011-068. PMID: 22614668. PMCID: PMC3721957.
35. Chen Q, Zhang H, Liu Y, Adams S, Eilken H, Stehling M, Corada M, Dejana E, Zhou B, Adams RH. 2016; Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nat Commun. 7:12422. DOI: 10.1038/ncomms12422. PMID: 27516371. PMCID: PMC4990645. PMID: 442c956f54bf4c6eba0ef2653c390245.

36. Pardali E, Sanchez-Duffhues G, Gomez-Puerto MC, Ten Dijke P. 2017; TGF-β-induced endothelial-mesenchymal transition in fibrotic diseases. Int J Mol Sci. 18:2157. DOI: 10.3390/ijms18102157. PMID: 29039786. PMCID: PMC5666838.

37. Wong WT, Cooke JP. 2016; Therapeutic transdifferentiation of human fibroblasts into endothelial cells using forced expression of lineage-specific transcription factors. J Tissue Eng. 7:2041731416628329. DOI: 10.1177/2041731416628329. PMID: 27081470. PMCID: PMC4820020.

38. Marzoog BA, Vlasova TI. 2021; Beta-cell autophagy under the scope of hypoglycemic drugs; possible mechanism as a novel therapeutic target. Obe Metab. 18:465–70. DOI: 10.14341/omet12778.

39. Berendam SJ, Koeppel AF, Godfrey NR, Rouhani SJ, Woods AN, Rodriguez AB, Peske JD, Cummings KL, Turner SD, Engelhard VH. 2019; Comparative transcriptomic analysis identifies a range of immunologically related functional elaborations of lymph node associated lymphatic and blood endothelial cells. Front Immunol. 10:816. DOI: 10.3389/fimmu.2019.00816. PMID: 31057546. PMCID: PMC6478037.

40. Keuschnigg J, Karinen S, Auvinen K, Irjala H, Mpindi JP, Kallioniemi O, Hautaniemi S, Jalkanen S, Salmi M. 2013; Plasticity of blood- and lymphatic endothelial cells and marker identification. PLoS One. 8:e74293. DOI: 10.1371/journal.pone.0074293. PMID: 24058540. PMCID: PMC3769239. PMID: aa5f6f4cdead469a84ca43ce114f6dd0.

41. Yang J, Zhang S, Zhang L, Xie X, Wang H, Jie Z, Gu M, Yang JY, Cheng X, Sun SC. 2019; Lymphatic endothelial cells regulate B-cell homing to lymph nodes via a NIK-dependent mechanism. Cell Mol Immunol. 16:165–77. DOI: 10.1038/cmi.2017.167. PMID: 29503445. PMCID: PMC6355805.

42. Russo E, Runge P, Jahromi NH, Naboth H, Landtwing A, Montecchi R, Leicht N, Hunter MC, Takai Y, Halin C. 2021; CD112 regulates angiogenesis and T cell entry into the spleen. Cells. 10:169. DOI: 10.3390/cells10010169. PMID: 33467729. PMCID: PMC7830896. PMID: 48c9bb8f0a614e5d888c34399f17462b.

43. Claro V, Ferro A. 2020; Netrin-1: focus on its role in cardiovascular physiology and atherosclerosis. JRSM Cardiovasc Dis. 9:2048004020959574. DOI: 10.1177/2048004020959574. PMID: 33282228. PMCID: PMC7691900. PMID: 6d1ae565a2334a41a52233a84b211e6e.

44. Dieterich LC, Tacconi C, Menzi F, Proulx ST, Kapaklikaya K, Hamada M, Takahashi S, Detmar M. 2020; Lymphatic MAFB regulates vascular patterning during developmental and pathological lymphangiogenesis. Angiogenesis. 23:411–23. DOI: 10.1007/s10456-020-09721-1. PMID: 32307629. PMCID: PMC7311381.

45. Jourde-Chiche N, Fakhouri F, Dou L, Bellien J, Burtey S, Frimat M, Jarrot PA, Kaplanski G, Le Quintrec M, Pernin V, Rigothier C, Sallée M, Fremeaux-Bacchi V, Guerrot D, Roumenina LT. 2019; Endothelium structure and function in kidney health and disease. Nat Rev Nephrol. 15:87–108. DOI: 10.1038/s41581-018-0098-z. PMID: 30607032.

46. Becker PW, Sacilotto N, Nornes S, Neal A, Thomas MO, Liu K, Preece C, Ratnayaka I, Davies B, Bou-Gharios G, De Val S. 2016; An intronic Flk1 enhancer directs arterial-specific expression via RBPJ-mediated venous repression. Arterioscler Thromb Vasc Biol. 36:1209–19. DOI: 10.1161/ATVBAHA.116.307517. PMID: 27079877. PMCID: PMC4894770.
47. Carman CV, Martinelli R. Bradshaw RA, Stahl PD, editors. 2016. Lymphocyte-endothelial interactions. Encyclopedia of Cell Biology. Elsevier;Waltham: p. 632–49. DOI: 10.1016/B978-0-12-394447-4.30095-5.

48. Ribatti D. Ribatti D, editor. 2017. The origins of lymphatic vessels: an historical review. Milestones in Immunology: Based on Collected Papers. Elsevier;London: p. 129–62. DOI: 10.1016/B978-0-12-811313-4.00010-3.
49. Motawe ZY, Abdelmaboud SS, Breslin JW. 2021; Involvement of sigma receptor-1 in lymphatic endothelial barrier integrity and bioenergetic regulation. Lymphat Res Biol. 19:231–9. DOI: 10.1089/lrb.2020.0060. PMID: 33226886. PMCID: PMC8220569.

50. Park HS, Kim SY. 2021; Endothelial cell senescence: a machine learning-based meta-analysis of transcriptomic studies. Ageing Res Rev. 65:101213. DOI: 10.1016/j.arr.2020.101213. PMID: 33189866.

51. Li C, Tan Y, Wu J, Ma Q, Bai S, Xia Z, Wan X, Liang J. 2020; Resveratrol improves Bnip3-related mitophagy and attenuates high-fat-induced endothelial dysfunction. Front Cell Dev Biol. 8:796. DOI: 10.3389/fcell.2020.00796. PMID: 32923443. PMCID: PMC7457020. PMID: 373f21a8c3fe47b692cf70671f1ac543.

52. Zhao YY, Gao XP, Zhao YD, Mirza MK, Frey RS, Kalinichenko VV, Wang IC, Costa RH, Malik AB. 2006; Endothelial cell-restricted disruption of FoxM1 impairs endothelial repair following LPS-induced vascular injury. J Clin Invest. 116:2333–43. DOI: 10.1172/JCI27154. PMID: 16955137. PMCID: PMC1555637.

53. Li J, Huang NF, Zou J, Laurent TJ, Lee JC, Okogbaa J, Cooke JP, Ding S. 2013; Conversion of human fibroblasts to functional endothelial cells by defined factors. Arterioscler Thromb Vasc Biol. 33:1366–75. DOI: 10.1161/ATVBAHA.112.301167. PMID: 23520160. PMCID: PMC3898631.

54. Huang X, Zhang X, Zhao DX, Yin J, Hu G, Evans CE, Zhao YY. 2019; Endothelial hypoxia-inducible factor-1α is required for vascular repair and resolution of inflammatory lung injury through forkhead box protein M1. Am J Pathol. 189:1664–79. DOI: 10.1016/j.ajpath.2019.04.014. PMID: 31121134. PMCID: PMC6680254.

55. Liu M, Zhang L, Marsboom G, Jambusaria A, Xiong S, Toth PT, Benevolenskaya EV, Rehman J, Malik AB. 2019; Sox17 is required for endothelial regeneration following inflammation-induced vascular injury. Nat Commun. 10:2126. DOI: 10.1038/s41467-019-10134-y. PMID: 31073164. PMCID: PMC6509327. PMID: aef5d536288346a9aed9848b8a1f6268.

56. McDonald AI, Shirali AS, Aragón R, Ma F, Hernandez G, Vaughn DA, Mack JJ, Lim TY, Sunshine H, Zhao P, Kalinichenko V, Hai T, Pelegrini M, Ardehali R, Iruela-Arispe ML. 2018; Endothelial regeneration of large vessels is a biphasic process driven by local cells with distinct proliferative capacities. Cell Stem Cell. 23:210–25.e6. DOI: 10.1016/j.stem.2018.07.011. PMID: 30075129. PMCID: PMC6178982.

57. Huang X, Dai Z, Cai L, Sun K, Cho J, Albertine KH, Malik AB, Schraufnagel DE, Zhao YY. 2016; Endothelial p110γPI3K mediates endothelial regeneration and vascular repair after inflammatory vascular injury. Circulation. 133:1093–103. DOI: 10.1161/CIRCULATIONAHA.115.020918. PMID: 26839042. PMCID: PMC4792690.

58. Imrie H, Viswambharan H, Sukumar P, Abbas A, Cubbon RM, Yuldasheva N, Gage M, Smith J, Galloway S, Skromna A, Rashid ST, Futers TS, Xuan S, Gatenby VK, Grant PJ, Channon KM, Beech DJ, Wheatcroft SB, Kearney MT. 2012; Novel role of the IGF-1 receptor in endothelial function and repair: studies in endothelium-targeted IGF-1 receptor transgenic mice. Diabetes. 61:2359–68. DOI: 10.2337/db11-1494. PMID: 22733797. PMCID: PMC3425420.
59. Hutter R, Carrick FE, Valdiviezo C, Wolinsky C, Rudge JS, Wiegand SJ, Fuster V, Badimon JJ, Sauter BV. 2004; Vascular endothelial growth factor regulates reendothelialization and neointima formation in a mouse model of arterial injury. Circulation. 110:2430–5. DOI: 10.1161/01.CIR.0000145120.37891.8A. PMID: 15477421.

60. Eyries M, Siegfried G, Ciumas M, Montagne K, Agrapart M, Lebrin F, Soubrier F. 2008; Hypoxia-induced apelin expression regulates endothelial cell proliferation and regenerative angiogenesis. Circ Res. 103:432–40. DOI: 10.1161/CIRCRESAHA.108.179333. PMID: 18617693.

61. Miyagawa K, Shi M, Chen PI, Hennigs JK, Zhao Z, Wang M, Li CG, Saito T, Taylor S, Sa S, Cao A, Wang L, Snyder MP, Rabinovitch M. 2019; Smooth muscle contact drives endothelial regeneration by BMPR2-Notch1-mediated metabolic and epigenetic changes. Circ Res. 124:211–24. DOI: 10.1161/CIRCRESAHA.118.313374. PMID: 30582451. PMCID: PMC6400637.

62. Tang X, Luo YX, Chen HZ, Liu DP. 2014; Mitochondria, endothelial cell function, and vascular diseases. Front Physiol. 5:175. DOI: 10.3389/fphys.2014.00175. PMID: 24834056. PMCID: PMC4018556.

63. Kumar V, Jurkunas UV. 2021; Mitochondrial dysfunction and mitophagy in Fuchs endothelial corneal dystrophy. Cells. 10:1888. DOI: 10.3390/cells10081888. PMID: 34440658. PMCID: PMC8392447. PMID: f86c6b57bb624613bd9804c11cdc89ba.

64. Lekakis J, Abraham P, Balbarini A, Blann A, Boulanger CM, Cockcroft J, Cosentino F, Deanfield J, Gallino A, Ikonomidis I, Kremastinos D, Landmesser U, Protogerou A, Stefanadis C, Tousoulis D, Vassalli G, Vink H, Werner N, Wilkinson I, Vlachopoulos C. 2011; Methods for evaluating endothelial function: a position statement from the European Society of Cardiology Working Group on Peripheral Circulation. Eur J Cardiovasc Prev Rehabil. 18:775–89. DOI: 10.1177/1741826711398179. PMID: 21450600.

65. Shao X, Lai D, Zhang L, Xu H. 2016; Induction of autophagy and apoptosis via PI3K/AKT/TOR pathways by azadirachtin A in Spodoptera litura cells. Sci Rep. 6:35482. DOI: 10.1038/srep35482. PMID: 27752103. PMCID: PMC5067515.

66. Chen Y, Li P, Peng Y, Xie X, Zhang Y, Jiang Y, Li T, Qin X, Li S, Yang H, Wu C, Zheng C, Zhu J, You F, Liu Y. 2021; Protective autophagy attenuates soft substrate-induced apoptosis through ROS/JNK signaling pathway in breast cancer cells. Free Radic Biol Med. 172:590–603. DOI: 10.1016/j.freeradbiomed.2021.07.005. PMID: 34242793.

67. He C, Zhu H, Li H, Zou MH, Xie Z. 2013; Dissociation of Bcl-2-Beclin1 complex by activated AMPK enhances cardiac autophagy and protects against cardiomyocyte apoptosis in diabetes. Diabetes. 62:1270–81. DOI: 10.2337/db12-0533. PMID: 23223177. PMCID: PMC3609561.

68. Martin-Sanchez D, Poveda J, Fontecha-Barriuso M, Ruiz-Andres O, Sanchez-Niño MD, Ruiz-Ortega M, Ortiz A, Sanz AB. 2018; Targeting of regulated necrosis in kidney disease. Nefrologia (Engl Ed). 38:125–35. DOI: 10.1016/j.nefroe.2018.02.004. PMID: 28647049.

Fig. 1
Two primary complexes are involved in autophagosome formation, the first include of III PI3K Vps34, Atg6/Beclin1, Atg14, and Vps15/p150.73, whereas the second include the serine/threonine kinase Atg1 signaling axis. Furthermore, autophagy is done in two phases; the first phase includes the formation of phagophores and the second phase includes elongation that involved ubiquitin-like conjugation pathways; Atg8/MAP-LC3/GABAP/GATE-16 and Atg12 systems, in the center of the figure. Inducing autophagy can be done in various ways, indicated by a plus sign. During the second phase, Atg8 protein undergoes cleavage for its carboxyl-terminal by cysteine protease Atg4. Then, Atg4 undergoes a long complexed signaling cascade involved activation of Atg4 by Atg7 and Atg3 that culminate in activation of Atg8 and it is binding with the LC3–II of the lysosome and formation of Atg8/LC3–II complex. The Atg8/LC3–II complex bind with autophagosome in a covalent bound unless Atg4 cleaves it to be recycled and degraded by the phagolysosome. Not necessarily, the presented modulators can target all these pathways and signaling steps.
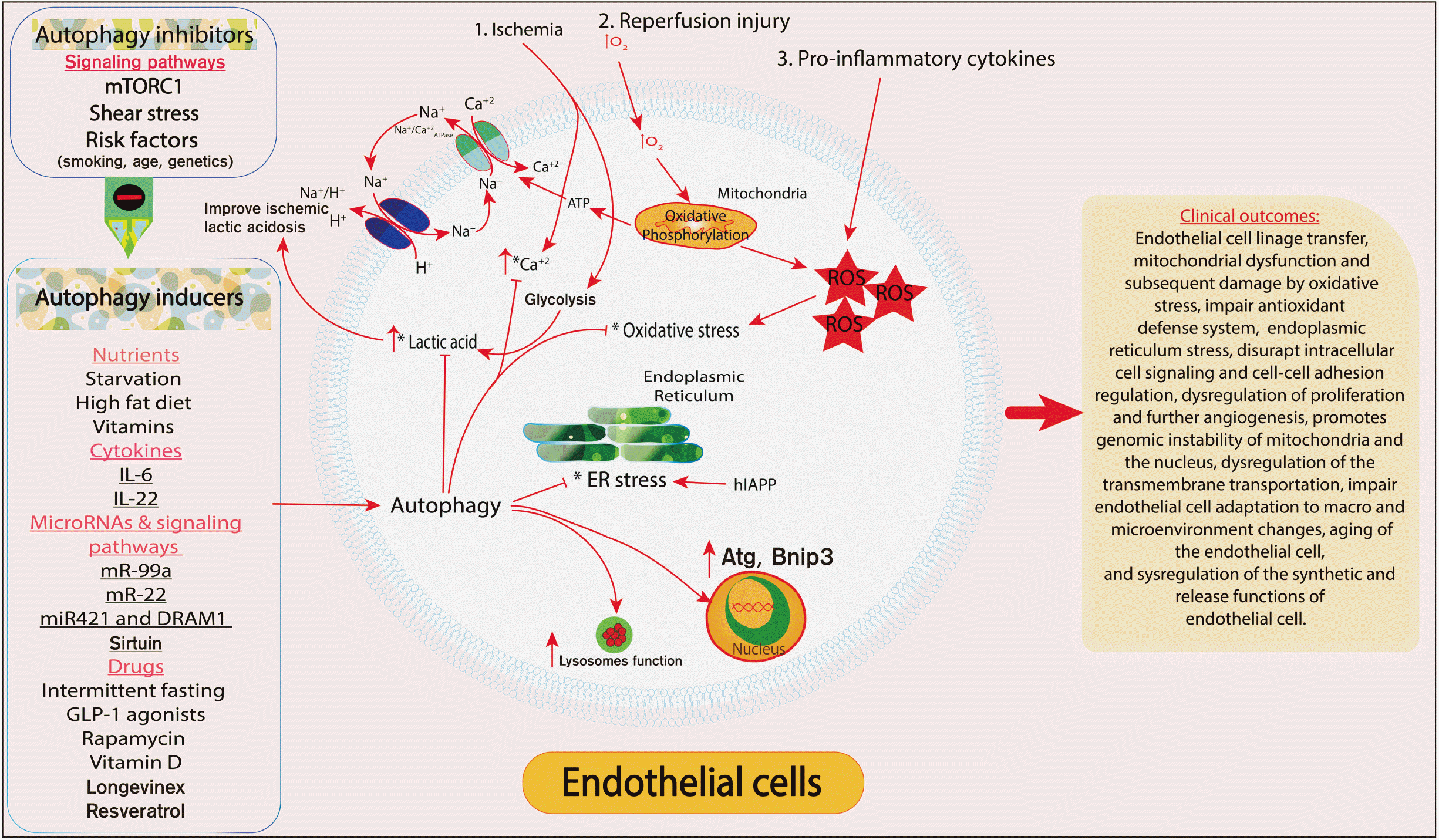
Fig. 2
Schematic presentation of the role of autophagy in endothelial cells. Autophagy prevents cell linage transfer, stabilizes mitochondrial function subsequent protection from oxidative stress, enhance cellular antioxidant defense system, prevents endoplasmic reticulum stress, maintains intracellular cell signaling, cell-cell adhesion regulation, enhances proliferation and further angiogenesis, promotes genomic stability of mitochondria and the nucleus, enhance transmembrane transportation, improves endothelial cell adaptation to macro and microenvironment changes, prevents endothelial cell aging, and improves the synthetic and release function of impaired endothelial cell.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download