Abstract
Background and Objectives
Lymphoblastoid cell lines (LCLs) deposited from disease-affected individuals could be a valuable donor cell source for generating disease-specific induced pluripotent stem cells (iPSCs). However, generation of iPSCs from the LCLs is still challenging, as yet no effective gene delivery strategy has been developed.
Methods and Results
Here, we reveal an effective gene delivery method specifically for LCLs. We found that LCLs appear to be refractory toward retroviral and lentiviral transduction. Consequently, lentiviral and retroviral transduction of OCT4, SOX2, KFL4 and c-MYC into LCLs does not elicit iPSC colony formation. Interestingly, however we found that transfection of oriP/EBNA-1-based episomal vectors by electroporation is an efficient gene delivery system into LCLs, enabling iPSC generation from LCLs. These iPSCs expressed pluripotency makers (OCT4, NANOG, SSEA4, SALL4) and could form embryoid bodies.
Induced pluripotent stem cells (iPSCs) can be generated from different types of somatic cells including fibroblasts and keratinocytes by ectopic expression of four reprogramming factors OCT4, SOX2, KLF4, and MYC (hereafter referred to as OSKM) (1). iPSCs hold great promise for regenerative medicine by virtue of their unlimited proliferation and multilineage differentiation capacity (1, 2). More specifically, iPSCs generated from isolated patient cells and their derivatives provide powerful in vitro models with which elucidation of cellular and molecular mechanisms underlying disease pathogenesis and development of new drugs to alleviate disease phenotypes are possible (3). Furthermore, genetically and phenotypically corrected patient-specific iPSCs can be used for preclinical and clinical trials to investigate their potential for replacing damaged cells/tissues (2).
Pelizaeus-Merzbacher disease (PMD) is an X-linked recessive hypomyelination disorder that is caused by mutations in the proteolipid protein 1 (PLP1) (4). PLP1 is specifically expressed in oligodendrocyte precursor cells (OPCs) and oligodendrocytes (OLs) and plays decisive roles in compaction, stabilization, and maintenance of myelin sheaths (5). Dysregulation of these processes that are elicited by mutations in PLP1 can lead to death of OLs, consequently resulting in the loss of myelin sheaths (6, 7). More than 100 different mutations in PLP1 have been reported in PMD, including duplications, point mutations and null mutations (4). Duplications of PLP1 are the most common mutations, which have been detected in over 60% of PMD patients. Missense and nonsense PLP1 mutations have been reported in over 20% PDM patients. Null mutations are relatively rare. Clinical symptoms (phenotypes) of PMD patients are diverse (4), but whether/how these phenotypic diversities between PMD patents are actually linked to different types of genotypic mutations remains unknown. Furthermore, cellular and molecular mechanisms underlying the pathogenesis of PMD are not yet fully defined. Hence, establishment of iPSC lines from individuals with mutations spanning genetic and clinical diversities of PMD may enhance our understanding of PMD pathogenesis and genotype-phenotype relationship.
Lymphoblastoid cell lines (LCLs) have been generated by infection of B cells from peripheral blood of healthy donors and patients with Epstein-Barr Virus (EBV) (8). These EBV-immortalized LCLs are a convenient and useful model to study a variety of human genetic disorders. In fact, they have been used for various biomedical studies including disease-related gene discovery and mutational analyses (9, 10). Importantly, over tens thousands of LCLs have been deposited internationally from a variety of individuals with various ethnic backgrounds and disease states (11-13). For instance, the Coriell Institute for Medical Re-search currently stores twenty-three LCLs which are deposited from ten PMD patients and thirteen related heal-thy family members. iPSCs generated from these LCLs and their derivatives (OPCs and OLs) could provide a valuable model for studying demyelination diseases and discovering therapeutic options for PMD.
In this study, we attempted to develop an efficient gene delivery method for LCLs. We found that electroporation is an efficient means to deliver genes into LCLs. With this strategy, we successfully generated PMD-specific iPSCs from LCLs. These iPSCs expressed pluripotency markers and could form embryoid bodies. Our findings show that electroporation is an effective method with which disease-specific iPSCs can be generated from LCLs.
Epstein-Barr virus immortalized lymphoblastoid cell lines (LCLs) were obtained from the Coriell Institute. The Coriell cell repository maintains the consent and privacy of the donor LCLs. LCLs were cultured in RPMI 1640 (Gibco) supplemented with 15% fetal bovine serum (FBS; Biochrom) and 2 mM L-glutamine (Sigma-Aldrich). CRL-2097 neonatal skin fibroblasts and HEK293 cells were purchased from ATCC and cultured in Dulbecco’s Modified Eagle’s medium high glucose (DMEM; Welgene) supplemented with 10% FBS, 1×nonessential amino acids (NEAAs; Sigma-Aldrich), 1×GlutaMAX (Gibco), and 1×penicillin/treptomycin (P/S; 100 U/ml each, Sigma-Aldrich). Human iPSCs were cultured on the Matrigel-coated plate with MEF-conditioned medium (MEF-CM). The MEF-CM medium was prepared as described previously (ref). The cells described in this study were cultured at 37℃ and 5% CO2 in a humidified Heraeus BB15 incubator (Thermo Fisher Scientific).
pLEX307-GFP and pCXLE-mCherry were cloned in house. pMXs-OCT4, pMXs-SOX2, pMXs-KLF4, pMXs-c-MYC, pMXs-GFP and pRRL-hOSKM-tdTomato were previously described (14-16). Other plasmids including pMD2.G (#12259), psPAX2 (#12260), pUMVC (#8449), pCMV-VSV-G (#8454), pCXLE-hOCT3/4 (#27076), pCXLE-hSK (#27078), pCXLE-hUL (#27080), and pCXLE-mp5 3DD (#41859) were purchased from Addgene. Plasmid DNA was isolated from E. coli and purified using Nucleo-Bond Xtra Kit (Macherey-Nagel).
To produce retroviruses, 4.5 μg of pMXs-GFP, 3 μg of pUMVC, and 1.5 μg of pCMV-VSV-G were transfected into HEK293 cells using 27 μl of Polyethylenimine (PEI; Polysciences) in 600 μl Opti-MEM (Invitrogen). At 48 h post-transfection, virus-containing supernatants were collected and filtered through a 0.45-μm PVDF filter (Mil-lipore), concentrated at 23,000 rpm for 2 h using an Optima XL-100 K Ultracentrifuge (Beckman), and resuspended in 1 ml of DMEM. Retroviral suspensions were stored at −80℃ until use. To produce lentiviruses, 1.5 μg of pMD2.G, 3 μg of psPAX2 and 4.5 μg of the pRRL-hOSKM-tdTomato were transfected into HEK293 cells using 27 μl of PEI in 600 μl of Opti-MEM. At 48 h post-tra-nsfection, virus-containing supernatants were collected and filtered through a 0.45-μm PVDF filter, concentrated at 23,000 rpm for 2 h using an Optima XL-100 K Ultrace-ntrifuge and resuspended in 1 ml of DMEM. Lentiviral suspensions were stored at −80℃ until use. The LCLs and CRL2097 fibroblasts were then infected with viruses twice over the course of three days in the presence of 8 μg/ml protamine sulfate (Sigma-Aldrich).
Sodium butyrate (NaB; Sigma-Aldrich) was used at a final concentration of 250 μM. SB431542 (purity: ≥98%; Cayman Chemical) was used at a final concentration of 2 μM. SGC0946 (purity: ≥98%; Cayman Chemical) was used at a final concentration of 3 μM.
1×106 of LCLs were electroporated with total 2 μg of pCXLE-hOCT3/4, pCXLE-hSK, pCXLE-hUL, and pCXLE-mp53DD (0.5 μg each) using the NeonTM Transfection System 10 μl Kit (MPK10096; Thermo Fisher Scientific). The transfected LCLs were then transferred to 35 mm dishes pre-coated with hESC-qualified Matrigel (BD Bios-ciences) and cultured in MEF-CM supplemented with NaB, SB431542, and SGC0946. The medium was changed every other day. Chemicals were added until colonies reached the size of over 200 μm in diameter.
The cells were mechanically or enzymatically disso-ciated and resuspended in 300 μl of 3% FBS/PBS. The cell suspension was transferred to 5 ml polystyrene round-bottom tubes through a cell strainer cap (Falcon) and subjected to flow cytometry analysis. The cells were then separated from debris and aggregates by forward scatter/side scatter (FSC/SSC) gating. Single cells were identified by plotting FSC area versus FSC width. Dead cells were excluded by staining with DAPI and gating on DAPI+ cells. Cells that had not been infected with viruses or had been electroporated with empty plasmids were used as negative controls for gating. Fluorescence was measured using a FACSCanto II (BD Biosciences) and flow cytometry data were analyzed using FlowJo software (Tree Star Inc.).
The cells were fixed with 10% formalin (Cellnest) for 20 mins, incubated with 0.1% Triton X-100/PBS for 30 mins, and blocked in 5% BSA/PBS for 1 h. The cells were incubated with appropriate primary antibodies overnight at 4℃. The primary antibodies used were following: anti-SSEA-4 (1:100, 90231; Millipore), anti-NANOG (1:1,000, 5232S, Cell Signaling Technology), anti-OCT4 (1:1,000, 5677S; Cell Signaling Technology), anti-SALL4 (1:1,000, ab29112; Abcam). The cells were then washed three times with PBS and incubated with appropriate fluorescently labeled Alexa-Fluor secondary antibodies (1:1,000, Invitrogen) for 1 hr. The cells were then washed three times with PBS, incubated with 0.5 μg/ml 4’,6-dia-midino-2-phenylindole (DAPI, Molecular Probes) for 10 mins, and washed once with PBS. The cells were then subjected to immunofluorescence microscopy analysis. Images were acquired using a fluorescence inverted microscope (IX71; Olympus) equipped with the CCD camera (DP30BW; Olympus) and analyzed with the DP-BSW (Olympus) and Fiji software (17).
Retroviral transduction of OSKM has been widely used for generation of iPSCs from different types of human cells (15, 18). Since retroviruses can only infect actively dividing cells (19), our first choice for gene delivery into LCLs was retroviral transduction. We transduced LCLs with retroviruses containing OSKM and cultured in MEF-CM medium supplemented with sodium butyrate (a HDAC inhibitor), SB431542 (a TGFb inhibitor) and SGC0946 (a DOT1L inhibitor) (Fig. 1A). These chemicals positively influence iPSC generation (20). Surprisingly, retroviral transduction of OSKM in LCLs did not yield any iPSC colonies in contrast to concurrently transduced fibroblasts (Fig. 1B). We then hypothesized that the failure of generating iPSCs from LCLs might be due the fact that retrovirus might not effectively transduce LCLs. To test this hypothesis, we transduced LCLs with retroviruses encoding green fluorescent protein (GFP) and evaluated GFP expression by a fluorescence microscope. At five days post-infection, we found that GFP was not expressed in retroviral GFP-transduced LCLs (Fig. 1C). Neither increased titers of GFP virus or longer culturing periods (days 7 and 9) changed GFP negativity (Fig. 1C). In contrast, GFP was expressed in retroviral GFP-transduced fibroblasts (Fig. 1D). Flow cytometry analysis further confirmed that GFP was not expressed in retrovirally GFP-transduced LCLs, in contrast to concurrently transduced fibroblasts which yield over 20% GFP+ cells at day 5 post-infection (Fig. 1E). Together, these data demonstrate a decisive difference in retroviral transduction between two cell types and show that retrovirus-mediated gene delivery is not effective for LCLs.
Lentiviruses can infect a wider range of cell types than retroviruses (21). We thus postulated that lentiviral transduction might be in fact an effective way to deliver genes into LCLs. To test this, we produced lentiviruses with a polycistronic lentiviral vector encoding OCT4, SOX2, KLF4, c-MYC, and tdTomato (hereafter referred as OSKMT) and transduced LCLs (Fig. 2A). In this system, tdTomato was linked to OSKM by an internal ribosomal entry site (IRES) and thus transgene expression can be monitored by tdTomato expression. Surprisingly, at forty-eight hours post-infection, we found that lentiviral OSKMT-transduced LCLs did not yield any tdTomato+ cells, in contrast to concurrently transduced fibroblasts which yielded over 70% tdTomato+ cells (Fig. 2B∼D). Consequently, le-ntiviral OSKMT-transduced LCLs failed to produce iPSC colonies, whereas concurrently transduced fibroblasts produced iPSC colonies (Fig. 2E). Together these data demonstrate lentiviral transduction is also not an effective way to deliver genes into LCLs and is not suitable for generating iPSCs from LCLs.
Beside retroviral and lentiviral transduction, electroporation is an efficient gene delivery method which can introduce genes into a wide range of cell types, especially hard-to-transfect cells, like primary cells and cells that are cultured in suspension (22, 23). Electroporation uses an electrical pulse to create temporary pores in cell membranes through which substances like plasmids can enter into the cells. Different cell types require distinct electroporation settings (i.e. pulse time, voltage strength) to achieve high viability and reproducible transfection efficiency (22, 23). To identify an optimal electroporation condition for LCLs, we electroporated LCLs with episomal vector encoding mCherry with different ranges of voltage stren-gth and pulse width (Fig. 3A). We selected an OriP/EBNA1-based episomal vector particularly as a vehicle to deliver mCherry into LCLs, since two elements (oriP and EBNA-1) allow for replication and long-term retainment of vectors in mammalian cells (24). At forty-eight hours post-transfection, mCherry expression was analyzed by a fluorescence microscope. Intriguingly, of six condition we tested, two conditions (20 ms/1,400 v, 30 ms/1,100 v) yie-lded over 20% mCherry+ cells (Fig. 3B and 3C). Of note, cell death rates increased with higher voltage conditions (20 width: 1,700 v, 30 width: 1,400 v, 40 width: 1,200 v), indicating that voltage strengths influence the cell viability (Fig. 3C). Together these data reveal defined electroporation conditions that enable efficient gene delivery into LCLs.
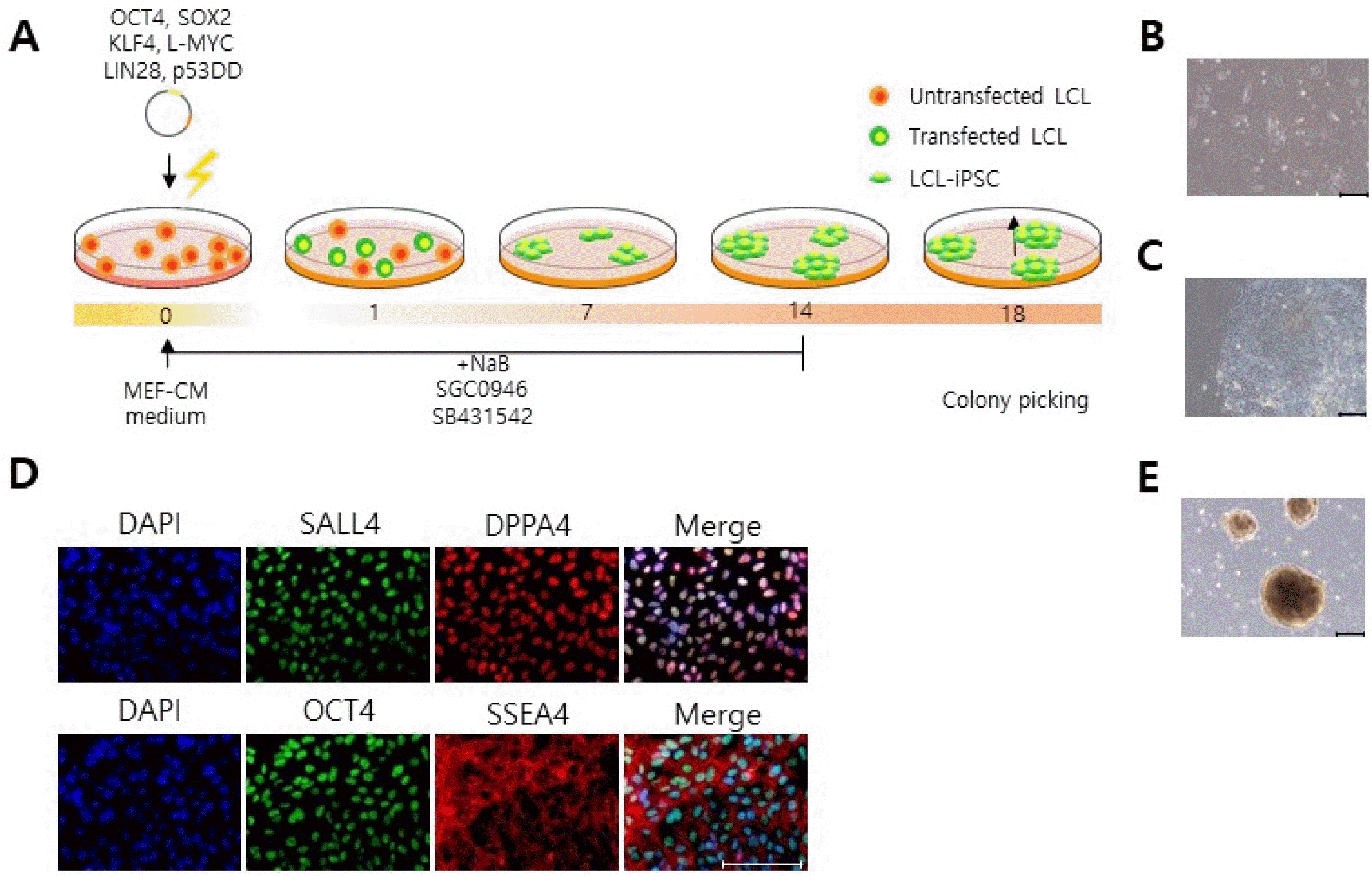
Having established that electroporation is a powerful and versatile method for delivery genes into LCLs, we next attempted to use this method to reprogram LCLs into iPSCs. For this, we electroporated LCLs, which were derived from a PDM patient, with episomal vectors encoding repro-graming factors [OCT4, SOX2, KLF4, L-MYC, LIN28 and p53DD (a dominant-negative inhibitor of p53 function)] and subsequently cultured them in MEF-CM medium supplemented with sodium butyrate, SB431542, and SGC0946 (Fig. 4A). At one week after transfection, we observed a number of small iPSC colonies firmly attached to the bottom of the culture plate (Fig. 4B). After the colonies reached over 800 μm in diameter (Fig. 4C), they were mechanically isolated and cultured independently. To confirm if the LCLs had transformed into iPSCs, we stained them with the number of pluripotency makers (Fig. 4D). We found that the LCL-derived iPSCs indeed expressed OCT4, SSEA4, DPPA4, and SALL4 (Fig. 4D). Further-more, they could form embryoid bodies when the cells were cultured in ultra-low attachment culture plates (Fig. 4E). Together these data demonstrate that electroporation of episomal vectors enables iPSC generation from LCLs.
Lymphoblastoid cell lines (LCLs) can grow indefinitely in culture and derive from B lymphocytes infected by Epstein-Barr virus (EBV) (8). Because of their non-adherent nature in culture, LCLs are relatively easy to handle by researchers. Over tens thousands of LCLs have been deposited from a variety of patients with various ethnic backgrounds and disease states (11-13). Disease-specific iPSCs generated from these LCLs and their derivatives would provide a powerful in vitro model for elucidating disease pathogenesis and developing new therapeutic options.
Although LCLs are actively dividing cells (8), we found that retroviral transduction could not infect LCLs. Why LCLs appear to be refractory towards retroviral transduction remains unknown. However, one could readily expect that certain retroviral receptors are not present in LCLs such that retroviruses cannot dock to the surface of LSCs to initiate infection. Thus, it will be interesting to determine whether ectopic expression of some or any retroviral receptors might render LCLs susceptible to retrovirus infection.
Lentiviruses are capable of infecting a wider range of cell type and also it can infect even non-dividing cells (21). As such, our observation that LCLs cannot be transduced by lentiviruses was unexpected. For lentivirus-mediated reprogramming, we used a polycistronic lentiviral vector encoding OSKMT (pRRL-OSKMT) (16). In this vector, a retroviral spleen focus-forming virus U3 (SFFV) promoter regulates OSKMT expression. This promoter is known to mediate efficient expression of genes in only certain types of cells, but not in all cell types (16). Furthermore, it has been shown that this promoter can be rapidly silenced in some cell types, like hematopoietic cells and pluripotent stem cells (16, 25). Thus, the failure of lentiviral transduction in LCLs might be due to the activity of SFFV promoter present in pRRL-OSKMT. We found that elongation factor 1 alpha (EF1α) promoter-derived lentiviral GFP was effectively expressed in LCLs, which supports this idea (Supplementary Fig. S1A∼C).
Given the tremendous potential of iPSCs for regene-rative medicine and disease modeling, generation of disease-specific iPSCs have become important (2). In this study, we generated PMD-specific iPSCs from LCLs which derived from a PMD patient. PMD is a demyelinating disease that eventually results in developmental delays and neurological dysfunction, and death (4). Current pharmaceutical treatments cannot cure PMD. With PMD-iPSCs and their differentiated derivatives (OPCs and OLs), future studies will aim to develop new therapeutic perspectives and investigate PMD pathogenesis.
Supplementary data including one figure can be found with this article online at https://doi.org/10.15283/ijsc22177.
References
1. Takahashi K, Yamanaka S. 2016; A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 17:183–193. DOI: 10.1038/nrm.2016.8. PMID: 26883003.
2. Yamanaka S. 2020; Pluripotent stem cell-based cell therapy-promise and challenges. Cell Stem Cell. 27:523–531. DOI: 10.1016/j.stem.2020.09.014. PMID: 33007237.
3. Inoue H, Nagata N, Kurokawa H, Yamanaka S. 2014; iPS cells: a game changer for future medicine. EMBO J. 33:409–417. DOI: 10.1002/embj.201387098. PMID: 24500035. PMCID: PMC3989624.
4. Osório MJ, Goldman SA. 2018; Neurogenetics of Pelizaeus-Merzbacher disease. Handb Clin Neurol. 148:701–722. DOI: 10.1016/B978-0-444-64076-5.00045-4. PMID: 29478609. PMCID: PMC8167836.
5. Elitt MS, Barbar L, Shick HE, Powers BE, Maeno-Hikichi Y, Madhavan M, Allan KC, Nawash BS, Gevorgyan AS, Hung S, Nevin ZS, Olsen HE, Hitomi M, Schlatzer DM, Zhao HT, Swayze A, LePage DF, Jiang W, Conlon RA, Rigo F, Tesar PJ. 2020; Suppression of proteolipid protein rescues Pelizaeus-Merzbacher disease. Nature. 585:397–403. DOI: 10.1038/s41586-020-2494-3. PMID: 32610343. PMCID: PMC7810164.
6. Nevin ZS, Factor DC, Karl RT, Douvaras P, Laukka J, Windrem MS, Goldman SA, Fossati V, Hobson GM, Tesar PJ. 2017; Modeling the mutational and phenotypic landscapes of Pelizaeus-Merzbacher disease with human iPSC-derived oligodendrocytes. Am J Hum Genet. 100:617–634. DOI: 10.1016/j.ajhg.2017.03.005. PMID: 28366443. PMCID: PMC5384098.
7. Nobuta H, Yang N, Ng YH, Marro SG, Sabeur K, Chavali M, Stockley JH, Killilea DW, Walter PB, Zhao C, Huie P Jr, Goldman SA, Kriegstein AR, Franklin RJM, Rowitch DH, Wernig M. 2019; Oligodendrocyte death in Pelizaeus-Merz-bacher disease is rescued by iron chelation. Cell Stem Cell. 25:531–541.e6. DOI: 10.1016/j.stem.2019.09.003. PMID: 31585094. PMCID: PMC8282124.
8. Omi N, Tokuda Y, Ikeda Y, Ueno M, Mori K, Sotozono C, Kinoshita S, Nakano M, Tashiro K. 2017; Efficient and reliable establishment of lymphoblastoid cell lines by Epstein-Barr virus transformation from a limited amount of peripheral blood. Sci Rep. 7:43833. DOI: 10.1038/srep43833. PMID: 28272413. PMCID: PMC5341036.
9. Lappalainen T, Sammeth M, Friedländer MR, Monlong J, Rivas MA, Gonzàlez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, Greger L, van Iterson M, Almlöf J, Ribeca P, Pulyakhina I, Esser D, Giger T, Tikhonov A, Sultan M, Bertier G, MacArthur DG, Lek M, Lizano E, Buermans HP, Padioleau I, Schwarzmayr T, Karlberg O, Ongen H, Kilpinen H, Beltran S, Gut M, Kahlem K, Amstislavskiy V, Stegle O, Pirinen M, Mont-gomery SB, Donnelly P, McCarthy MI, Flicek P, Strom TM, Lehrach H, Schreiber S, Sudbrak R, Carracedo A, Antonarakis SE, Häsler R, Syvänen AC, van Ommen GJ, Brazma A, Meitinger T, Rosenstiel P, Guigó R, Gut IG, Estivill X, Dermitzakis ET. 't Hoen PA. Geuvadis Consortium. 2013; Transcriptome and genome sequencing uncovers functional variation in hu-mans. Nature. 501:506–511. DOI: 10.1038/nature12531. PMID: 24037378. PMCID: PMC3918453.
10. Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, Johnson R, Segrè AV, Djebali S, Niarchou A, Wright FA, Lappalainen T, Calvo M, Getz G, Dermitzakis ET, Ardlie KG, Guigó R. GTEx Consortium. 2015; Human genomics. The human transcriptome across tissues and individuals. Science. 348:660–665. DOI: 10.1126/science.aaa0355. PMID: 25954002. PMCID: PMC4547472.
11. ENCODE Project Consortium. 2012; An integrated encyclopedia of DNA elements in the human genome. Nature. 489:57–74. DOI: 10.1038/nature11247. PMID: 22955616. PMCID: PMC3439153.
12. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. 1000 Genomes Project Consortium. 2015; A global reference for human genetic variation. Nature. 526:68–74. DOI: 10.1038/nature15393. PMID: 26432245. PMCID: PMC4750478.
13. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Gibbs RA, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Dermitzakis E, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Bonnen PE, Gibbs RA, Gonzaga-Jauregui C, Keinan A, Price AL, Yu F, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Schaffner SF, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Pollack S, Price AL, Schaffner SF, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. International HapMap 3 Consortium. 2010; Integrating common and rare genetic variation in diverse human populations. Nature. 467:52–58. DOI: 10.1038/nature09298. PMID: 20811451. PMCID: PMC3173859.
14. Kim KP, Li C, Bunina D, Jeong HW, Ghelman J, Yoon J, Shin B, Park H, Han DW, Zaugg JB, Kim J, Kuhlmann T, Adams RH, Noh KM, Goldman SA, Schöler HR. 2021; Donor cell memory confers a metastable state of directly converted cells. Cell Stem Cell. 28:1291–1306.e10. DOI: 10.1016/j.stem.2021.02.023. PMID: 33848472.
15. Kim KP, Wu Y, Yoon J, Adachi K, Wu G, Velychko S, MacCarthy CM, Shin B, Röpke A, Arauzo-Bravo MJ, Stehling M, Han DW, Gao Y, Kim J, Gao S, Schöler HR. 2020; Reprogramming competence of OCT factors is determined by transactivation domains. Sci Adv. 6:eaaz7364. DOI: 10.1126/sciadv.aaz7364. PMID: 32917606. PMCID: PMC7467702.
16. Warlich E, Kuehle J, Cantz T, Brugman MH, Maetzig T, Galla M, Filipczyk AA, Halle S, Klump H, Schöler HR, Baum C, Schroeder T, Schambach A. 2011; Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 19:782–789. DOI: 10.1038/mt.2010.314. PMID: 21285961. PMCID: PMC3070104.
17. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012; Fiji: an open-source platform for biological-image analysis. Nat Methods. 9:676–682. DOI: 10.1038/nmeth.2019. PMID: 22743772. PMCID: PMC3855844.
18. Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilić J, Pekarik V, Tiscornia G, Edel M, Boué S, Izpisúa Belmonte JC. 2008; Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 26:1276–1284. DOI: 10.1038/nbt.1503. PMID: 18931654.
19. Miller DG, Adam MA, Miller AD. 1990; Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol Cell Biol. 10:4239–4242. Erratum in: Mol Cell Biol 1992;12:433. DOI: 10.1128/MCB.10.8.4239. PMID: 2370865. PMCID: PMC360961.
20. Kim KP, Choi J, Yoon J, Bruder JM, Shin B, Kim J, Arauzo-Bravo MJ, Han D, Wu G, Han DW, Kim J, Cramer P, Schöler HR. 2021; Permissive epigenomes endow reprogramming competence to transcriptional regulators. Nat Chem Biol. 17:47–56. DOI: 10.1038/s41589-020-0618-6. PMID: 32807969.
21. Buchschacher GL Jr, Wong-Staal F. 2000; Development of lentiviral vectors for gene therapy for human diseases. Blood. 95:2499–2504. DOI: 10.1182/blood.V95.8.2499.008k35_2499_2504. PMID: 10753827.
22. Harris E, Elmer JJ. 2021; Optimization of electroporation and other non-viral gene delivery strategies for T cells. Biotech-nol Prog. 37:e3066. DOI: 10.1002/btpr.3066. PMID: 32808434.
23. Jordan ET, Collins M, Terefe J, Ugozzoli L, Rubio T. 2008; Optimizing electroporation conditions in primary and other difficult-to-transfect cells. J Biomol Tech. 19:328–334. PMID: 19183796. PMCID: PMC2628074.
24. Hammerschmidt W, Sugden B. 2013; Replication of Epstein-Barr viral DNA. Cold Spring Harb Perspect Biol. 5:a013029. DOI: 10.1101/cshperspect.a013029. PMID: 23284049. PMCID: PMC3579399.
25. Herbst F, Ball CR, Tuorto F, Nowrouzi A, Wang W, Zavidij O, Dieter SM, Fessler S, van der Hoeven F, Kloz U, Lyko F, Schmidt M, von Kalle C, Glimm H. 2012; Extensive methylation of promoter sequences silences lentiviral transgene expression during stem cell differentiation in vivo. Mol Ther. 20:1014–1021. DOI: 10.1038/mt.2012.46. PMID: 22434137. PMCID: PMC3345972.
Fig. 1
LCLs are refractory to retroviral transduction. (A) Schematic representation of retroviral transduction procedure. (B) Quantification of iPSC colonies that had emerged from retroviral OSKM-transduced LCLs and fibroblasts. Data are presented as mean±SEM (n=3). (C) No GFP+ cells were observed in retroviral GFP-transduced LCLs, as determined by a fluorescent microscope. 30 μl and 50 μl of GFP virus were used for transduction. scale bar, 100 μm. (D) GFP+ cells were observed in retroviral GFP-transduced fibroblasts, as determined by a fluorescent microscope. 50 μl of GFP virus were used for transduction. scale bar, 100 μm. (E) Flow cytometry analysis of retroviral OSKM-transduced LCLs and fibroblasts. Non-infected cells were used for gating.
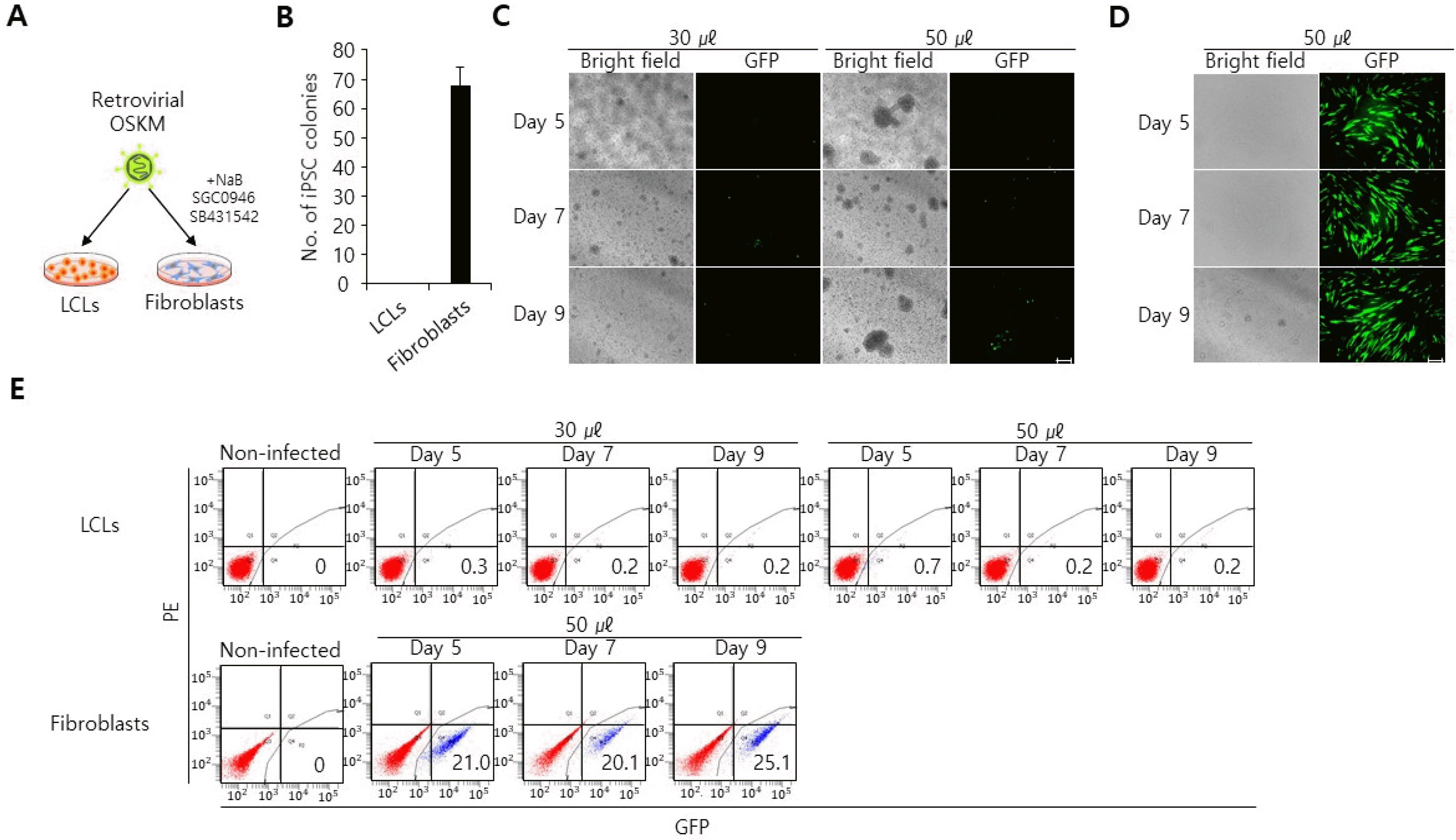
Fig. 2
LCLs are refractory to lentiviral transduction. (A) Schematic representation of lentiviral transduction procedure. (B) No tdTomato+ cells were observed in lentiviral OSKMT-transduced LCLs, as determined by a fluorescent microscope. 50 μl and 100 μl of tdTomato virus were used for transduction. scale bar, 100 μm. (C) tdTomato+ cells were observed in lentiviral OSKMT-transduced fibroblasts, as determined by a fluorescent microscope. 30 μl of GFP virus was used for transduction. scale bar, 100 μm. (D) Flow cytometry analysis of lentiviral OSKMT-transduced LCLs and fibroblasts. Non-infected cells were used for gating. (E) Quantification of iPSC colonies that had emerged from retroviral OSKM-transduced LCLs and fibroblasts. Data are presented as mean±SEM (n=3).
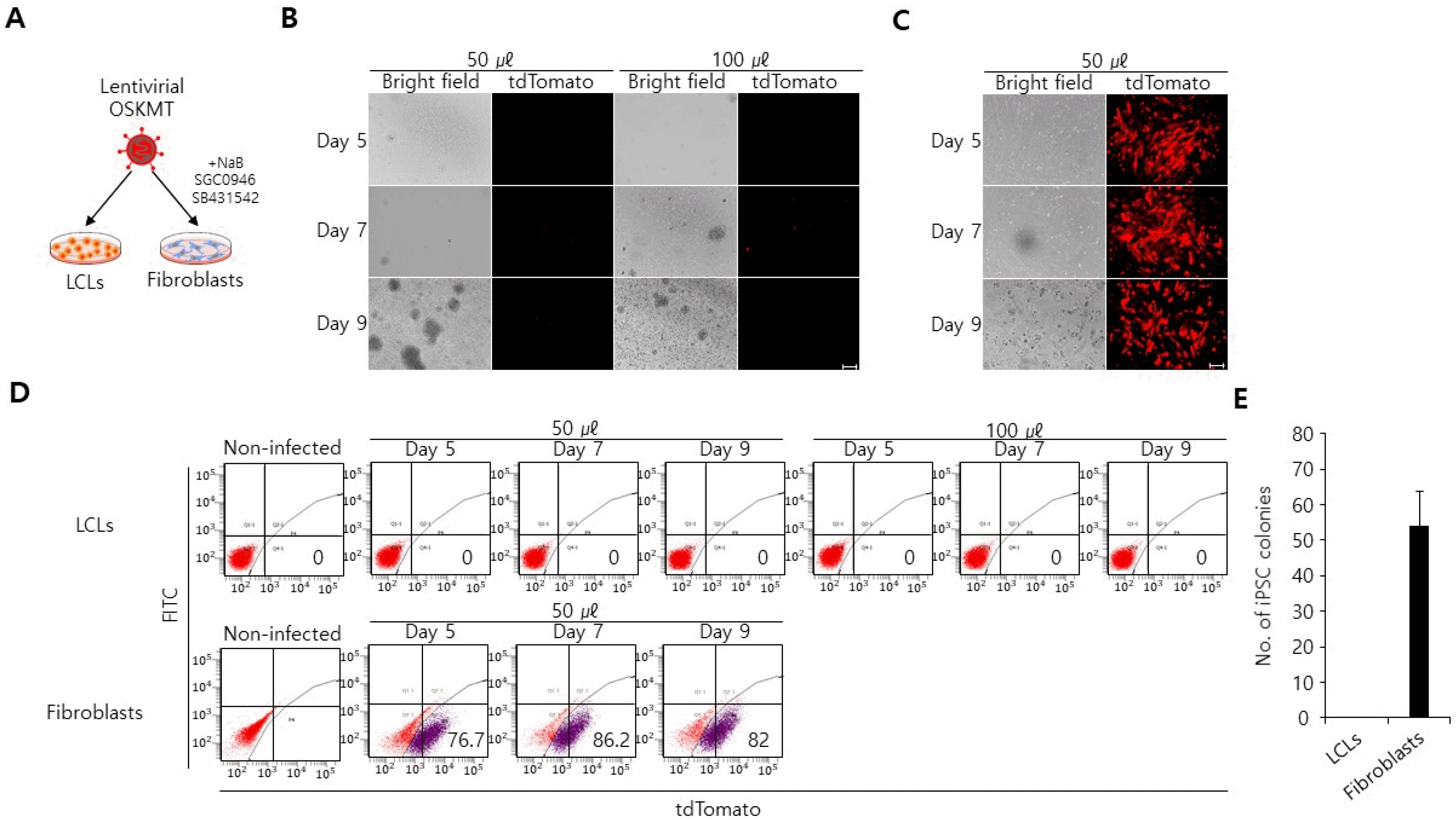
Fig. 3
Electroporation is an efficient method to deliver genes into LCLs. (A) Schematic representation of electroporation procedure. (B) Different rates of mCherry+ cells were detected in LCLs that had electroporated with different rages of voltage strength and pulse width. scale bar, 100 μm. (C) Flow cytometry analysis of LCLs that had electroporated with different rages of voltage strength and pulse width. The non-transfected cells were used for gating.
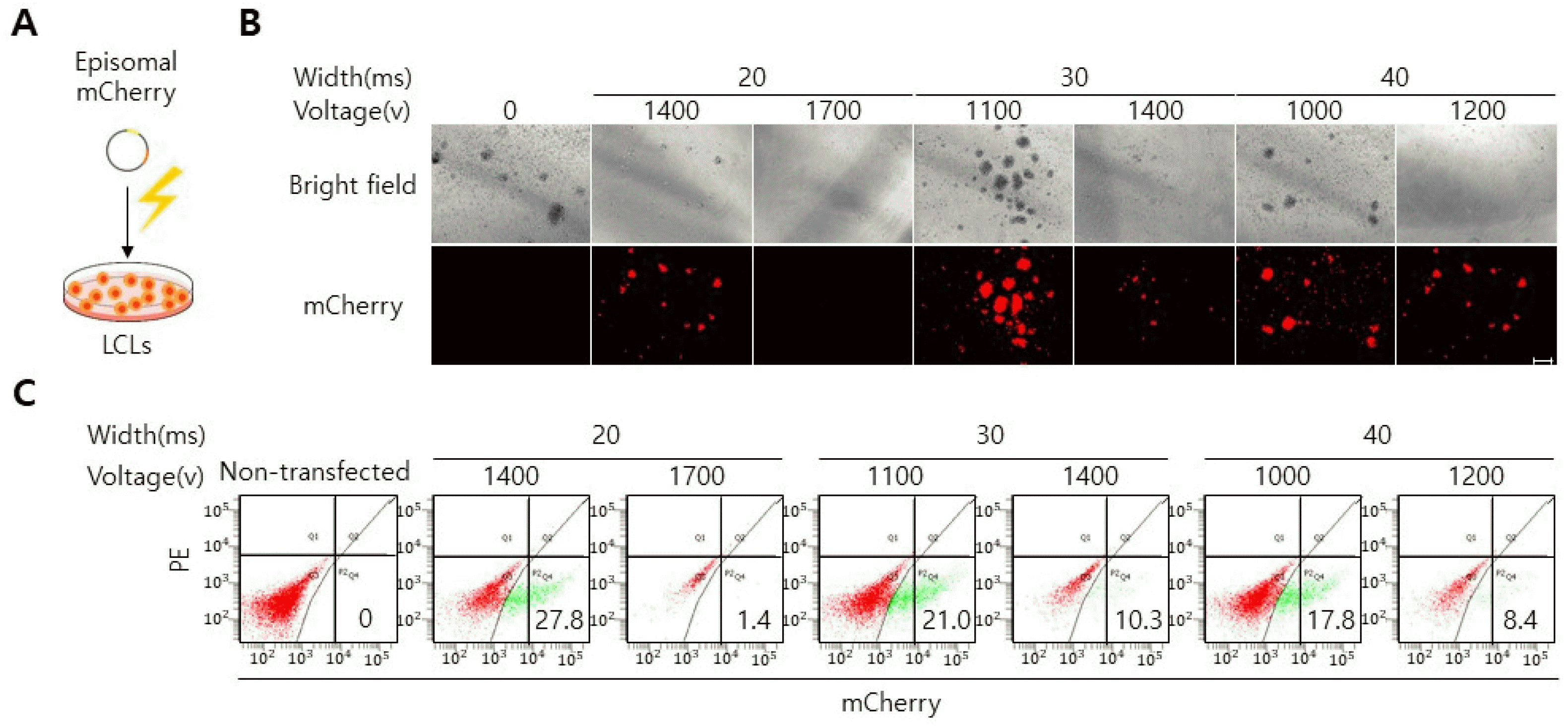
Fig. 4
Reprograming of LCLs into iPSCs by electroporation. (A) Schematic representation of reprogramming procedure. We electroporated LCLs with episomal vector encoding reprograming factors and cultured them in MEF-CM medium supplemented with sodium butyrate, SB431542 and SGC0946. These compounds were supplied to the medium for the first 14 days. (B) At one week post-transfection, small iPSC colonies were emerged in the plate. scale bar, 200 μm. (C) At three weeks post-transfection, the iPSC colonies appear large enough for picking. (D) Expression of pluripotency makers (SALL4, DPPA4, OCT4, and SSEA4) in iPSCs. DAPI was used for staining cell nuclei. scale bar, 100 μm. (E) iPSCs formed embryoid bodies. scale bar, 200 μm.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download