Abstract
Background and Objectives
Spermatogonial stem cells (SSCs) are the most primitive cells in spermatogenesis and are the only adult stem cells capable of passing on the genome of a given species to the next generation. SSCs are the only adult stem cells known to exhibit high Oct4 expression and can be induced to self-reprogram into pluripotent cells depending on culture conditions. Epigenetic modulation is well known to be involved in the induction of pluripotency of somatic cells. However, epigenetic modulation in self-reprogramming of SSCs into pluripotent cells has not been studied.
Methods and Results
In this study, we examined the involvement of epigenetic modulation by assessing whether selfreprogramming of SSCs is enhanced by treatment with epigenetic modulators. We found that second-generation selective class I HDAC inhibitors increased SSC reprogramming efficiency, whereas non-selective HDAC inhibitors had no effect.
Conclusions
We showed that pluripotent stem cells derived from adult SSCs by treatment with small molecules with epigenetic modulator functions exhibit pluripotency in vitro and in vivo. Our results suggest that the mechanism of SSC reprogramming by epigenetic modulator can be used for important applications in epigenetic reprogramming research.
Pluripotency can be acquired by conversion into pluripotent stem cells (PSCs) following overexpression of specific transcription factors, which are key determinants of cell type (1). Forced ectopic expression of pluripotency-associated gene cocktails such as Oct4, Sox2, c-Myc, and Klf4 can reprogram somatic cells into PSCs (1, 2). Based on these reprogramming techniques, recent studies have focused on the molecular mechanism of reprogramming of somatic cells into PSCs. Epigenetic modifications such as DNA methylation and histone modifications are known to be key factors inducing pluripotency (3, 4). Gene expression in somatic and differentiated cells is maintained by characteristic patterns of DNA methylation and histone modifications, and successful reprogramming should re-establish all of these epigenetic modifications to a pluripotent state at the whole-genome level (5).
Spermatogonial stem cells (SSCs), unipotent germ cells programmed for spermatogenesis, are the most important cells in spermatogenesis (6) and are responsible for passing on genetic information to the next generation. SSCs are the only adult cell type expressing Oct4, which is essential for reprogramming somatic cells into induced PSCs (7-10). Our previous study reported that SSCs can be reprogrammed into germline-derived PSCs (gPSCs) and can be converted to a pluripotent state in vitro without the presence of exogenous factors in defined culture conditions with feeder cells (11). However, in these conditions, the involvement of feeder cells in epigenetic modification during reprogramming cannot be excluded. Therefore, a method for inducing pluripotency in feeder-free conditions was needed, and we later developed a feeder-free culture system, thus excluding their effect (12). In this system, the mechanism of SSC self-reprogramming can be studied because only homogeneous SSCs are used without the intervention of feeder cells.
In chemical reprogramming that reprogramming method excluding transgene, small molecules with epigenetic modification functions are used for induction of pluripotency in somatic cells. Since pluripotency induction is impossible without such small molecules, epigenetic modification appears to be essential for pluripotency acquisition (13). However, the mechanism of epigenetic modification related to self-reprogramming of SSCs and reprogramming of somatic cells has not yet been elucidated. In this study, we screened small molecules that modulate various epigenetic modifications such as DNA methyla-tion and histone deacetylases (HDACs) and used them to investigate the involvement of epigenetic modulation in the self-reprogramming of SSCs on the basis of the existing feeder-free reprogramming induction method.
Establishment of SSCs from Oct4-GFP/LacZ transgenic mice (C57BL/6 background) and Oct4-GFP transgenic mice (C57BL/6 background) was described previously (9, 11, 14) SSCs, gPSCs, feeder-free system for derivation of gPS cells (FF-gPSCs), and ESCs were generated and characterized in previous studies and were cultured according to protocols presented previously (9, 12, 15).
For screening of small molecules inducing pluripotency, SSCs were seeded in Matrigel (Corning, Manassas, VA, USA)-coated 96-well plates. After 1 day, the Screen-Well Epigenetics library (Enzo Life Sciences, Doral, FL, USA) and 7 molecules (16) were each added to the culture medium (Supplementary Table S1). The library and the 7 molecules were dissolved in DMSO (Sigma-Aldrich, Saint Louis, MO, USA).
For pluripotency induction SSCs were seed at a density of 1×105 cells per well in 24-well plates coated with Matrigel and cultured in expansion medium (9). The next day, medium was replaced with a medium containing 5 μM CI-994, 0.3 μM mocetinostat, or 1 μM entinostat and was changed every day for 4 days. Then the medium containing the three small molecules was replaced with expansion medium, which was changed every 2 days until Oct4-GFP colonies were formed.
SSCs were plated at 2.5×105 cells per well in 24-well plates or 5×105 cells per well in 12-well plates and cultured to observe gPSC conversion. After 4 weeks, GFP-positive colonies were counted to calculate self-reprogramming efficiency.
Total RNA was isolated by using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Total RNA (500 ng) was reverse-transcribed by using the OmniscriptRT Kit (Qiagen) in a total volume of 20 μl. PCR analysis was performed with gene-specific primers and Takara Ex Taq DNA polymerase (TaKaRa Bio Inc., Kusatsu, Shiga, Japan) according to the manufacturer’s instructions. The PCR conditions were as follows: 32 cycles at 94℃ for 30 s, 50∼65℃ for 30 s, and 72℃ for 30 s. The RT-PCR products were analyzed by electrophoresis in 1% agarose gels. Primer sequences are listed in Supplementary Table S2.
The microarray study was carried out using Illumina Mouse Ref-8v2.0 Expression BeadChips (Illumina Inc., San Diego, CA, USA) as previously described (9, 11, 14). Quality-checked cDNA samples were hybridized for 18 h with the BeadChips, washed, stained, and scanned following the guidelines and using materials and instrumentation supplied or suggested by the manufacturer.
SSCs were blocked with 0.5% BSA (Sigma-Aldrich) in PBS and stained with Alexa FluorⓇ 647 Mouse Anti-SSEA-1 (BD Biosciences, Franklin Lakes, NJ, USA) for 30 min at room temperature (RT). Samples were analyzed using FACSAria IIu (BD Biosciences) and FlowJo v10 software (BD Biosciences).
Cells were fixed in 4% paraformaldehyde (Sigma-Aldrich) for 15 min at RT, washed 3 times with DPBS, and then incubated in DPBS containing 0.5% Triton X-100 (Sigma-Aldrich), 1% BSA fraction V (Sigma-Aldrich), and 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) for 10 min at RT. The cells were briefly rinsed with DPBS and incubated with primary antibodies overnight at 4℃ or for 1 h at RT. The cells were then washed with 0.5% BSA (Sigma-Aldrich) in PBS and incubated with secondary antibodies (anti-mouse IgG or anti-mouse IgM; 1:200; R&D Systems Inc., Minnesota, USA) for 1 h at RT. Nuclei were stained with 4’-6-diamidino-2-phenylindole (Dapi; Sigma-Aldrich). Information on antibodies is provided in Supplementary Table S3.
Cells were washed with PBS, and genomic DNA was isolated using the Total DNA Extraction kit (Intron, Liberty Lake, WA, USA) according to the manufacturer’s protocol. Genomic DNA was treated with Epi-Tech Bisulfite (Qiagen) according to the manufacturer’s recommendations and used for PCR amplification. The PCR products were cloned using the PCR Cloning kit (Qiagen). Primer sequences are listed in Supplementary Table S4.
To differentiate CI-gPSCs into three germ layers, previously described protocols (17, 18) were applied to embryoid bodies derived from gPSCs. Embryoid bodies were attached to a gelatin-coated plate and cultured in MEF medium until beating cells were formed. MEF medium was Dulbecco’s modified Eagle’s medium (DMEM) low-glucose (Welgene, Gyeongsan, Korea) with the following supplements: 10% FBS (Gibco), 50 μM β-mercaptoethanol (Gibco), 1×penicillin–streptomycin (Welgene), and 1×minimal essential medium non-essential amino acids (Gibco).
CI-gPSCs were aggregated with denuded post-compacted 8-cell-stage embryos retrieved from mice (C57BL/6 albino) at 2.5 days post coitum (dpc). Clumps of loosely connected gPSCs (10∼20 cells) after short trypsin treatment were selected and transferred into microdrops of KSOM mouse embryo medium containing 10% FBS (Gibco) under mineral oil. Each clump was placed in a depression in the microdrop. Batches of 30 to 40 embryos were briefly incubated with acidified Tyrode’s solution until the zona pellucida was degraded. A single embryo was placed into each clump. All aggregates were cultured at 37℃ in a humidified atmosphere containing of 5% CO2. After 24 h, the majority of the aggregates had formed blastocysts. Approximately 11 to 14 aggregated embryos were transferred into the uterine horn of 2.5-dpc pseudopregnant mouse.
To induce pluripotency in SSCs by epigenetic regulation, SSCs were treated with of Screen-Well Epigenetics library or 7 molecules at 1∼10 μM (16) and small molecules involved in reprogramming were selected according to the intensity of Oct4-GFP. The Oct4-GFP-positive gPSC population increased in CI-994, a HDAC class I inhibitor, -treatedSSCs (Supplementary Fig. S1). To determine the optimal concentration, SSCs were treated with CI-994 at 0.1∼100 μM to find the concentration at which cell death did not occur even with continuous exposure (Supplementary Fig. S2). Treatment for 4 days at a highest concentration of 5 μM did not induce cell death, and GFP-positive colonies appeared 8 days after removal the small molecules (Fig. 1A and 1B). Oct4-GFP-positive CI-gPSCs had grown similar morphology as mESCs in feeder-free culture (Fig. 1C). Positivity for alkaline phosphatase, SSEA1, and Nanog indicated that CI-gPSCs expressed pluripotency markers (Fig. 1C). To verify the reprogramming efficiency of CI-994 under the optimized conditions, we compared Oct4-GFP-positive colony formation with or without CI-994 treatment (Fig. 1D). We observed that the number of Oct4-GFP-positive colonies in CI-994 was higher than control group. These results suggest that CI-994, induces and promotes self-reprogramming of SSCs.
RT-PCR results indicated that CI-gPSCs expressed the pluripotency-associated genes Oct4, Nanog, Esg1, Fgf4, Cripto, Rex1, Eras, and Utf1 (Fig. 2A). Scatter plots of microarray analysis demonstrated the difference in gene expression patterns between CI-gPSCs and SSCs and similarity between CI-gPSCs and ESCs (Fig. 2B). Hierar-chical clustering and heatmap analysis revealed that global gene expression of CI-gPSCs was more similar to that of gPSCs and ESCs than to that of FF-gPSCs, SSCs, and MEFs (Fig. 2C). Bisulfite sequencing analysis showed that Oct4 and Nanog were hypomethylated in ESCs, gPSCs, FF-PSCs, and CI-PSCs, but were hypermethylated in SSCs (Fig. 2D). Taken together, these results demonstrate that the cellular and molecular characteristics of CI-gPSCs were very similar to those of ESCs and other gPSCs.
We carried out in vitro and in vivo differentiation to confirm the pluripotency of CI-gPSCs. We investigated differentiation of CI-gPSCs in embryoid bodies into three germ layers and observed differentiated cells positive for AFP, an endoderm lineage marker α-SMA, a mesenchymal cell marker; and Tuj1, a neuronal cell marker (Fig. 3A∼C). The presence of the three germ layers proved that CI-gPSCs could differentiate in vitro. To confirm the in vivo differentiation potential of CI-gPSCs, we transplanted them into immunodeficient mice to generate teratomas. Within 4 weeks of transplantation, teratomas formed. In the H&E analysis, staining of a teratoma section revealed that tissues contained all three germ layers, confirming that CI-gPSCs differentiated into the endoderm (gland), mesoderm (muscle), and ectoderm (neural rosette) (Fig. 3D∼F). The presence of teratomas containing three germ layers showed that CI-gPSCs could differentiate in vivo. To further assess the pluripotency of CI-gPSCs, we investigated chimera formation. The chimera analysis showed the contribution of CI-gPSCs to all three germ layers—endoderm (gut), mesoderm (skeletal muscle), ectoderm (skin)—and to germ cells, as confirmed by the presence of Oct4-GFP-positive cells in the fetal gonads of chimeric embryos (Fig. 3G∼I). These results clearly show that CI-gPSCs not only differentiate into the three germ layers, but also have pluripotency to contribute to the germline.
The DNA methylation patterns of differentially methylated regions (DMRs) are known not to change even after SSCs are converted to gPSCs (9). Therefore, we sought to confirm that methylation of DMRs of the paternally imprinted gene H19 and maternally imprinted gene Snrpn were maintained in the same CI-gPSCs as gPSCs and feeder free derived gPSCs generated previously (19). Bisulfite sequencing analysis revealed that H19 was hypermethylated, whereas Snrpn was hypomethylated (Supplementary Fig. S3) in CI-gPSCs.
CI-994 belong to class I HDAC inhibitors (Supplementary Table S5). To test whether various HDAC inhibitors would affect reprogramming, were carried out SSC reprogramming experiments using HDAC inhibitors of various classes. Oct4-GFP colonies were appear after 4 days of treatment with 0.3 μM mocetinostat or 1 μM entinostat, which are also class I HDAC inhibitors, followed by a week of incubation without HDAC inhibitors (Fig. 4A and 4B), whereas Oct4-GFP was not observed after treatment with inhibitors of other HDAC classes such as valproic acid (data not shown). After treatment of SSCs with CI-994, mocetinostat, and entinostat the number of colonies was approximately 4.5, 11.6, and 2.2 times that of untreated SSCs, respectively (Fig. 4C). These results indicate that class I HDAC could act as a barrier in SSC reprogramming and their inhibition by small molecules such as mocetinostat improves the efficiency of reprogramming.
SSCs are the only adult stem cells expressing Oct4 capable of self-reprogramming through pluripotency induction in defined culture conditions only without introducing specific genes or treatment with small molecules that are capable of epigenetic control for pluripotency induction (9, 11, 14). We tried to increase the efficiency of converting these SSCs into gPSCs through chemical screening. Although a close relation between somatic cell reprogramming and epigenetic modulation is known, studies on epigenetic modulation related to self-reprogramming of SSCs are still insufficient. In this study, we screened various epigenetic modulators to test whether epigenetic modulation is also related to the induction of pluripotency in SSCs, and found through the use of an Oct4-GFP reporting cell line that CI-994, a selective inhibitor of class I HDAC, can convert SSCs to gPSCs by further increasing the expression of endogenous Oct4. This suggests that the expression of endogenous Oct4 in the early culture stage may be related to the self-reprogramming of SSCs.
Studies aimed at improving the efficiency of Oct4 in reprogramming suggested that, since specific epigenetic modifications act as a barrier to reprogramming, removing these epigenetic modifications through genetic inactivation or chemical inhibition using small molecules can activate the expression of endogenous Oct4 to enhance reprogramming efficiency (20). These findings are consistent with our study, in which the expression of Oct4 was increased by removing the barrier to reprogramming using small molecules, thereby facilitating self-reprogramming.
In previous studies, several strategies have been developed to improve the efficiency of reprogramming using small molecules such as HDAC inhibitors (21). In many studies, transcription factors that enable induce pluripotency were replaced with compounds. Valproic acid (VPA) reprogrammed somatic cells with Oct4 and Sox2 by replacing c-Myc, and this combination increased induced pluripotent stem cells (iPSCs) colony numbers by 50-fold (22, 23). Human fetal fibroblasts were reprogrammed to pluripotency in culture supplemented with the HDAC inhibitors 5-azacytidine (5, 22), RG108 (24), TSA, and SAHA (22, 23). These findings imply that inhibition of a HDAC has a significant impact on the efficiency of reprogramming and are in part consistent with the results of our study, which found improved conversion of SSC-derived PSCs due to increased Oct4 expression by the HDAC inhibitors CI-994, mocetinostat, and entinostat.
However, not all of the small molecules tested showed the same effect in our study. VPA, a comprehensive epigenetic modulator, was not effective (data not shown). Although most studies on the effects of epigenetic modifications on reprogramming efficiency used somatic cells, our study is unique in that it used germ cells. Considering that pluripotency could not be induced in cancer cells (16, 25) even using same method that induce pluirpotency from somatic cells, the difference in the effects of these small molecules appears to be due to the cell type-dependent. Our study suggests that different epigenetic modification mechanisms may need to be engaged in different cells to induce pluripotency, as such mechanisms differ in different cells.
Although TSA and SAHA selectively inhibit class I HDAC, like the chemicals we found, they did not show any effect in our study. These results seem to require more detailed HDAC control. Inhibition of specific HDACs is known to increase reprogramming efficiency (26). TSA inhibits HDAC 1, 3, 4, 6, 10, and SAHA inhibits 1, 3. Among the compounds that showed an effect on SSCs, CI-994 inhibits 1, 2, 3, 8, mocetinostat inhibits 2, 3, 11, and entinostat inhibits 4, 6, 8, and 10. Therefore, the mechanism of acquiring pluripotency in SSCs appears to be related to the inhibition of HDAC 2 and 8, which belong to class I HDAC. The reports that selective inhibition of HDAC 8 increases the expression of the pluripotency marker genes Oct4, Nanog, and Sox2 and that selective inhibition of HDAC 2 increases that of Oct4 and Nanog are support the correlation between inhibition of HDAC 2 and 8 and pluripotency (26, 27). However, as mentioned above, pluripotency induction mechanisms differ depending on the cell type, our hypothesis that HDAC 2 and 8 relate with pluripotency may be a mechanism that is specific to SSCs, which already express Oct4, unlike somatic cells.
HDACs are classified into four classes according to structure, enzyme function, intracellular localization, and expression pattern (28, 29); among them, Class I, which is selectively inhibited by small molecules that were effective in our study, is closely related to development (30). Oct4, a gene essential for pluripotency induction, has functions related to early embryonic development, germ cell maintenance, stem cell pluripotency, and cell reprogramming (31-35). From these and our findings, it can be inferred that class I HDAC is closely related to Oct4 expression and reprogramming, and inhibition of HDAC 2 and 8 of this class may play a major role in inducing pluripotency. However, further studies are needed to understand the link between class I HDAC in general or specifically HDAC 2 and 8, and Oct4 in the self-reprogramming of SSCs.
In this study, we found that class I HDAC inhibitors are highly efficient in SSC reprogramming, suggesting that class I HDACs play an important role in the reprogramming of SSCs. This result suggests that epigenetic modulation is critical for induction of pluripotency in SSCs and specific modulation of HDACs is involved in self-reprogramming process in SSCs, which can help to identify the unique reprogramming mechanism of SSCs in future.
Supplementary data including five tables and three figures can be found with this article online at https://doi.org/10.15283/ijsc22110.
References
1. Takahashi K, Yamanaka S. 2006; Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 126:663–676. DOI: 10.1016/j.cell.2006.07.024. PMID: 16904174.
2. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007; Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 131:861–872. DOI: 10.1016/j.cell.2007.11.019. PMID: 18035408.
3. Bernstein BE, Meissner A, Lander ES. 2007; The mammalian epigenome. Cell. 128:669–681. DOI: 10.1016/j.cell.2007.01.033. PMID: 17320505.
4. Cedar H, Bergman Y. 2009; Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 10:295–304. DOI: 10.1038/nrg2540. PMID: 19308066.
5. Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. 2008; Dissecting direct reprogramming through integrative genomic analysis. Nature. 454:49–55. Erratum in: Nature 2008;454:794. DOI: 10.1038/nature07056. PMID: 18509334. PMCID: PMC2754827.
6. Tegelenbosch RA, de Rooij DG. 1993; A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res. 290:193–200. DOI: 10.1016/0027-5107(93)90159-D. PMID: 7694110.
7. Dann CT, Alvarado AL, Molyneux LA, Denard BS, Garbers DL, Porteus MH. 2008; Spermatogonial stem cell self-renewal requires OCT4, a factor downregulated during retinoic acid-induced differentiation. Stem Cells. 26:2928–2937. DOI: 10.1634/stemcells.2008-0134. PMID: 18719224.
8. Kanatsu-Shinohara M, Morimoto T, Toyokuni S, Shinohara T. 2004; Regulation of mouse spermatogonial stem cell self-rene-wing division by the pituitary gland. Biol Reprod. 70:1731–1737. DOI: 10.1095/biolreprod.103.025668. PMID: 14766726.
9. Ko K, Tapia N, Wu G, Kim JB, Bravo MJ, Sasse P, Glaser T, Ruau D, Han DW, Greber B, Hausdörfer K, Sebastiano V, Stehling M, Fleischmann BK, Brüstle O, Zenke M, Schöler HR. 2009; Induction of pluripotency in adult unipotent germline stem cells. Cell Stem Cell. 5:87–96. DOI: 10.1016/j.stem.2009.05.025. PMID: 19570517.
10. Pesce M, Wang X, Wolgemuth DJ, Schöler H. 1998; Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech Dev. 71:89–98. DOI: 10.1016/S0925-4773(98)00002-1. PMID: 9507072.
11. Ko K, Araúzo-Bravo MJ, Kim J, Stehling M, Schöler HR. 2010; Conversion of adult mouse unipotent germline stem cells into pluripotent stem cells. Nat Protoc. 5:921–928. DOI: 10.1038/nprot.2010.44. PMID: 20431537.
12. Lee SW, Wu G, Choi NY, Lee HJ, Bang JS, Lee Y, Lee M, Ko K, Schöler HR, Ko K. 2018; Self-reprogramming of spermatogonial stem cells into pluripotent stem cells without microenvironment of feeder cells. Mol Cells. 41:631–638. DOI: 10.14348/molcells.2018.2294. PMID: 29991673. PMCID: PMC6078851.
13. Bang JS, Choi NY, Lee M, Ko K, Park YS, Ko K. 2019; Reprogramming of cancer cells into induced pluripotent stem cells questioned. Int J Stem Cells. 12:430–439. DOI: 10.15283/ijsc19067. PMID: 31474029. PMCID: PMC6881048.
14. Ko K, Wu G, Araúzo-Bravo MJ, Kim J, Francine J, Greber B, Mühlisch J, Joo JY, Sabour D, Frühwald MC, Tapia N, Schöler HR. 2012; Autologous pluripotent stem cells generated from adult mouse testicular biopsy. Stem Cell Rev Rep. 8:435–444. DOI: 10.1007/s12015-011-9307-x. PMID: 21858421.
15. Choi NY, Park YS, Ryu JS, Lee HJ, Araúzo-Bravo MJ, Ko K, Han DW, Schöler HR, Ko K. 2014; A novel feeder-free culture system for expansion of mouse spermatogonial stem cells. Mol Cells. 37:473–479. DOI: 10.14348/molcells.2014.0080. PMID: 24854861. PMCID: PMC4086341.
16. Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, Ge J, Xu J, Zhang Q, Zhao Y, Deng H. 2013; Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 341:651–654. DOI: 10.1126/science.1239278. PMID: 23868920.
17. Brüstle O, Jones KN, Learish RD, Karram K, Choudhary K, Wiestler OD, Duncan ID, McKay RD. 1999; Embryonic stem cell-derived glial precursors: a source of myelinating transplants. Science. 285:754–756. DOI: 10.1126/science.285.5428.754. PMID: 10427001.
18. Igelmund P, Fleischmann BK, Fischer IR, Soest J, Gryshchenko O, Böhm-Pinger MM, Sauer H, Liu Q, Hescheler J. 1999; Action potential propagation failures in long-term recor-dings from embryonic stem cell-derived cardiomyocytes in tissue culture. Pflugers Arch. 437:669–679. DOI: 10.1007/s004240050831. PMID: 10087143.
19. Lee HJ, Choi NY, Lee SW, Ko K, Hwang TS, Han DW, Lim J, Schöler HR, Ko K. 2016; Epigenetic alteration of imprinted genes during neural differentiation of germline-derived pluripotent stem cells. Epigenetics. 11:177–183. DOI: 10.1080/15592294.2016.1146852. PMID: 26962997. PMCID: PMC4854545.
20. Kim KP, Han DW, Kim J, Schöler HR. 2021; Biological importance of OCT transcription factors in reprogramming and development. Exp Mol Med. 53:1018–1028. DOI: 10.1038/s12276-021-00637-4. PMID: 34117345. PMCID: PMC8257633.
21. Federation AJ, Bradner JE, Meissner A. 2014; The use of small molecules in somatic-cell reprogramming. Trends Cell Biol. 24:179–187. DOI: 10.1016/j.tcb.2013.09.011. PMID: 24183602. PMCID: PMC3943685.
22. Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. 2008; Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 26:795–797. DOI: 10.1038/nbt1418. PMID: 18568017. PMCID: PMC6334647.
23. Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. 2008; Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 26:1269–1275. DOI: 10.1038/nbt.1502. PMID: 18849973.
24. Shi Y, Desponts C, Do JT, Hahm HS, Schöler HR, Ding S. 2008; Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 3:568–574. DOI: 10.1016/j.stem.2008.10.004. PMID: 18983970.
25. Zhao Y, Zhao T, Guan J, Zhang X, Fu Y, Ye J, Zhu J, Meng G, Ge J, Yang S, Cheng L, Du Y, Zhao C, Wang T, Su L, Yang W, Deng H. 2015; A XEN-like state bridges somatic cells to pluripotency during chemical reprogramming. Cell. 163:1678–1691. DOI: 10.1016/j.cell.2015.11.017. PMID: 26686652.
26. Staszkiewicz J, Power RA, Harkins LL, Barnes CW, Strickler KL, Rim JS, Bondioli KR, Eilersten KJ. 2013; Silencing histone deacetylase-specific isoforms enhances expression of pluripotency genes in bovine fibroblasts. Cell Reprogram. 15:397–404. DOI: 10.1089/cell.2013.0026. PMID: 24020699. PMCID: PMC3787336.
27. Saha A, Pandian GN, Sato S, Taniguchi J, Hashiya K, Bando T, Sugiyama H. 2013; Synthesis and biological evaluation of a targeted DNA-binding transcriptional activator with HDAC8 inhibitory activity. Bioorg Med Chem. 21:4201–4209. DOI: 10.1016/j.bmc.2013.05.002. PMID: 23719282.
28. de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. 2003; Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 370(Pt 3):737–749. DOI: 10.1042/bj20021321. PMID: 12429021. PMCID: PMC1223209.
29. Gregoretti IV, Lee YM, Goodson HV. 2004; Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 338:17–31. DOI: 10.1016/j.jmb.2004.02.006. PMID: 15050820.
30. Haberland M, Montgomery RL, Olson EN. 2009; The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 10:32–42. DOI: 10.1038/nrg2485. PMID: 19065135. PMCID: PMC3215088.
31. Kang J, Shakya A, Tantin D. 2009; Stem cells, stress, metabolism and cancer: a drama in two Octs. Trends Biochem Sci. 34:491–499. DOI: 10.1016/j.tibs.2009.06.003. PMID: 19733480.
32. Pan GJ, Chang ZY, Schöler HR, Pei D. 2002; Stem cell pluripotency and transcription factor Oct4. Cell Res. 12:321–329. DOI: 10.1038/sj.cr.7290134. PMID: 12528890.
33. Shi G, Jin Y. 2010; Role of Oct4 in maintaining and regaining stem cell pluripotency. Stem Cell Res Ther. 1:39. DOI: 10.1186/scrt39. PMID: 21156086. PMCID: PMC3025441.
34. Tantin D. 2013; Oct transcription factors in development and stem cells: insights and mechanisms. Development. 140:2857–2866. DOI: 10.1242/dev.095927. PMID: 23821033. PMCID: PMC3699277.
35. Wu G, Schöler HR. 2014; Role of Oct4 in the early embryo development. Cell Regen. 3:7. DOI: 10.1186/2045-9769-3-7. PMID: 25408886. PMCID: PMC4230828.
Fig. 1
Pluripotency induction into gPSCs using small molecule CI-994. (A) Schematic of pluripotency induction into GFP-positive gPSCs using CI-994 treatment. (B) Time frame to convert SSCs to gPSCs. (C) Phase contrast and positive for alkialine phosphatase (AP), SSEA-1, and Nanog of CI-gPSCs. (D) Number of Oct4-GFP-positive gPSC colonies depending on CI-994 treatment. Scale bars: 200 μm (B) and 100 μm (C).
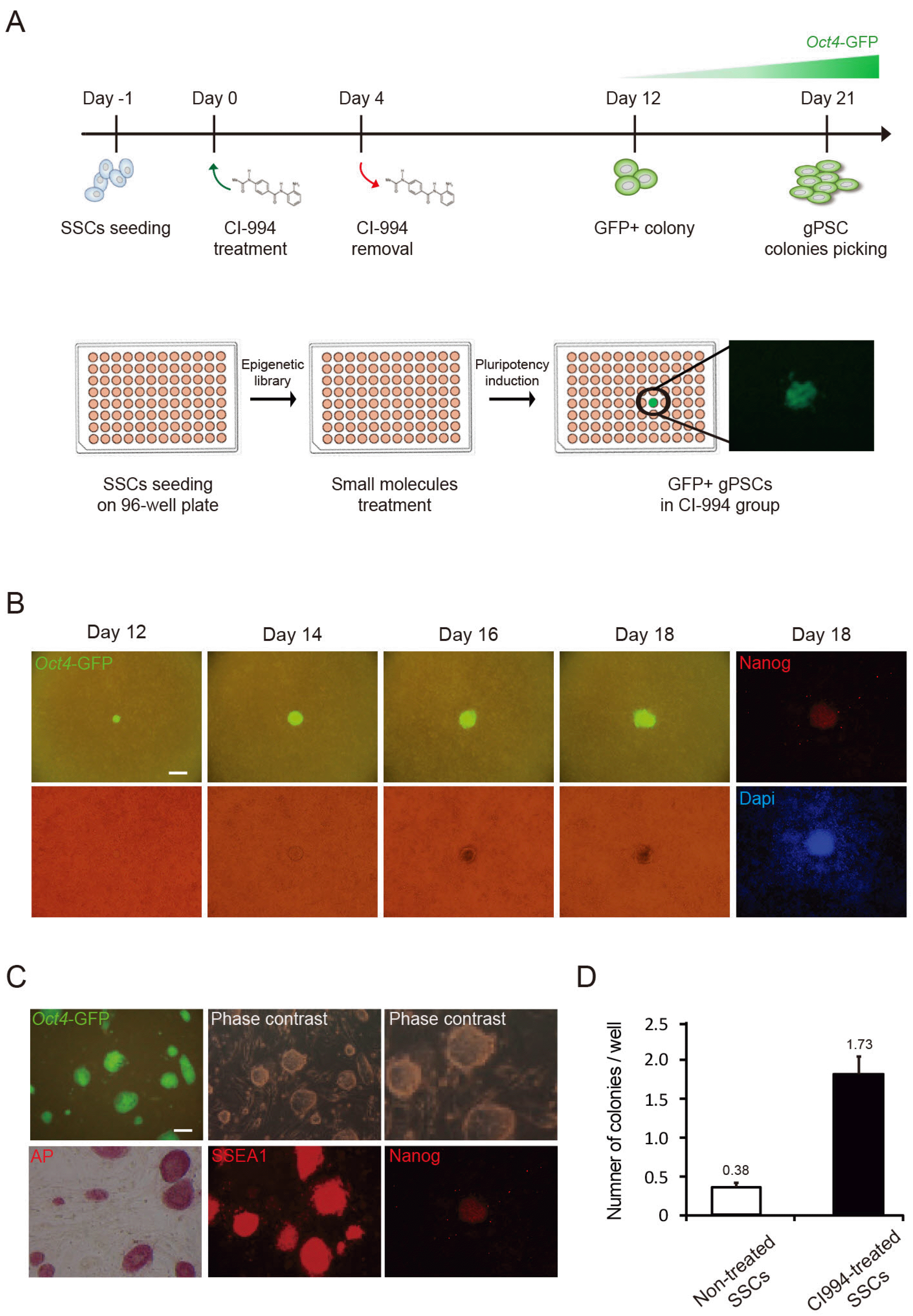
Fig. 2
Cellular and molecular characterization of CI-gPSCs. (A) RT-PCR analysis of pluripotency marker genes in ESCs, SSCs, gPSCs, FF-gPSCs (clone 1∼2), CI-gPSCs (clone 1∼3), and MEFs. β-actin was used as a loading control. (B) Comparison of global gene expression between CI-gPSCs and SSCs (left), and between CI-gPSCs and ESCs (right). (C) Hierarchical clustering and a heatmap analysis of MEFs, SSCs, FF-gPSCs (clone 1 and 6), CI-gPSCs (clone 1, 2, and 3), gPSCs, and ESCs. (D) Bisulfite genomic sequencing of promoter regions of Oct4 and Nanog in SSCs, gPSCs, FF-gPSCs, CI-gPSCs, and ESCs. Open and filled circles indicate unmethylated and methylated CpGs, respectively.
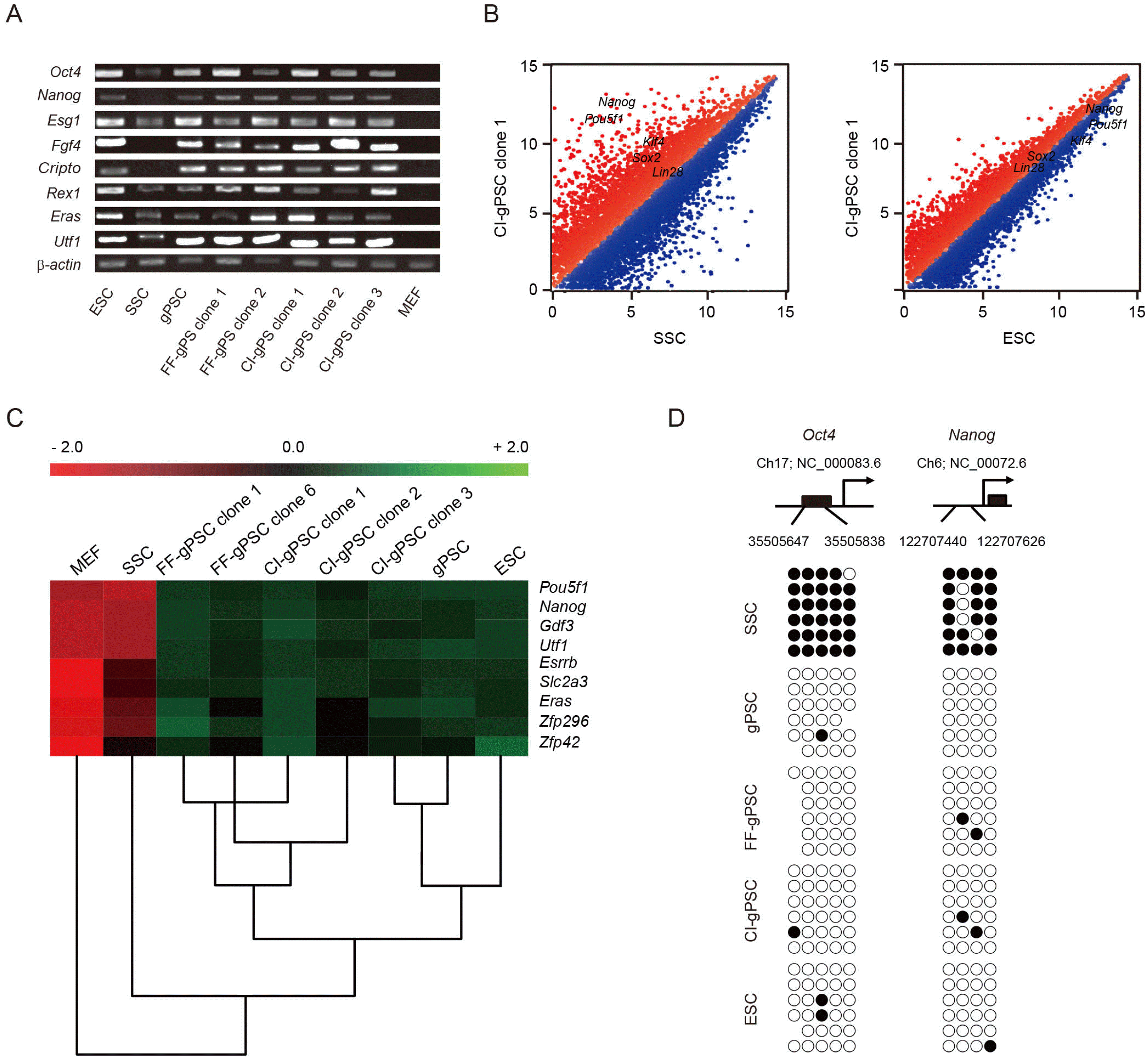
Fig. 3
Pluripotency of CI-gPSCs. (A∼C) In vitro differentiation into three germ layers. Stained cells positive in AFP, a-SMA, and Tuj1. (D∼F) H&E staining of teratoma-derived tissues. (G) Staining for β-galactosidase of chimeric mouse. (H∼I) Fetal gonads from embryo. White arrows indicate Oct4-GFP-positive SSCs in the gonad. Scale bars: 25 μm (A∼G), 100 μm (D∼F).
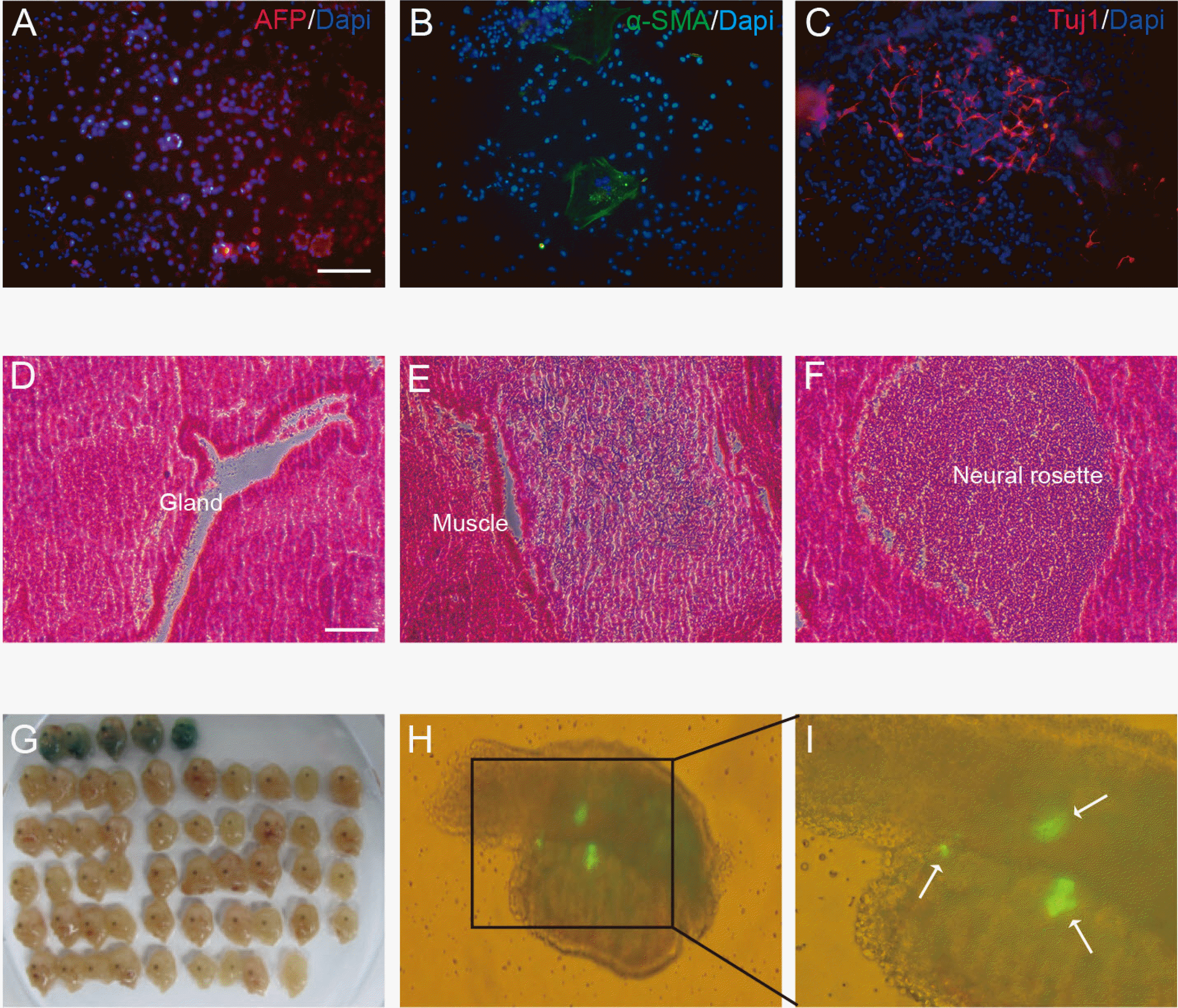
Fig. 4
Conversion of SSCs into gPSCs using Class I HDAC inhibitors mocetinostat and entinostat. (A) Experimental scheme for comparison of self-reprogramming efficiency of SSCs by HDAC inhibitor types. (B) Phase contrast photomicrograph and fluorescence images of Oct4-GFP in 0.5 μM mocetinostat and 1 μM entinostat treated SSCs. (C) Number of GFP-positive gPSC colonies depending on class I HDAC inhibitor type. Scale bars: 100 μm.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download