Dear Editor,
Point-of-care nucleic acid testing is essential for the rapid detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in emergent patients [1]. However, certain commercial PCR assays have low sensitivity for genetic variants, e.g., in the nucleocapsid (N) and envelope (E) genes [2, 3]. Variants such as T29194C, C29197T, and C29200T [4, 5] impair N2 detection by the Xpert Xpress SARS-CoV-2 assay (Cepheid, Sunnyvale, CA, USA), a commercial rapid PCR test with a turn-around time of 1 hour. In Korea, Xpert detection in N2 G29179T variant cases has resulted in high threshold cycle (Ct) values [6], which may lead to false-negative diagnoses and inadequate medical care for critically ill patients [7].
We report a case of N2-positive but E-negative detection by the Xpert assay in a patient pre-diagnosed as having coronavirus disease (COVID-19). The Institutional Review Board of Asan Medical Center, Seoul, Korea waived the requirement for review of the study and the need to obtain informed consent (2022-1362).
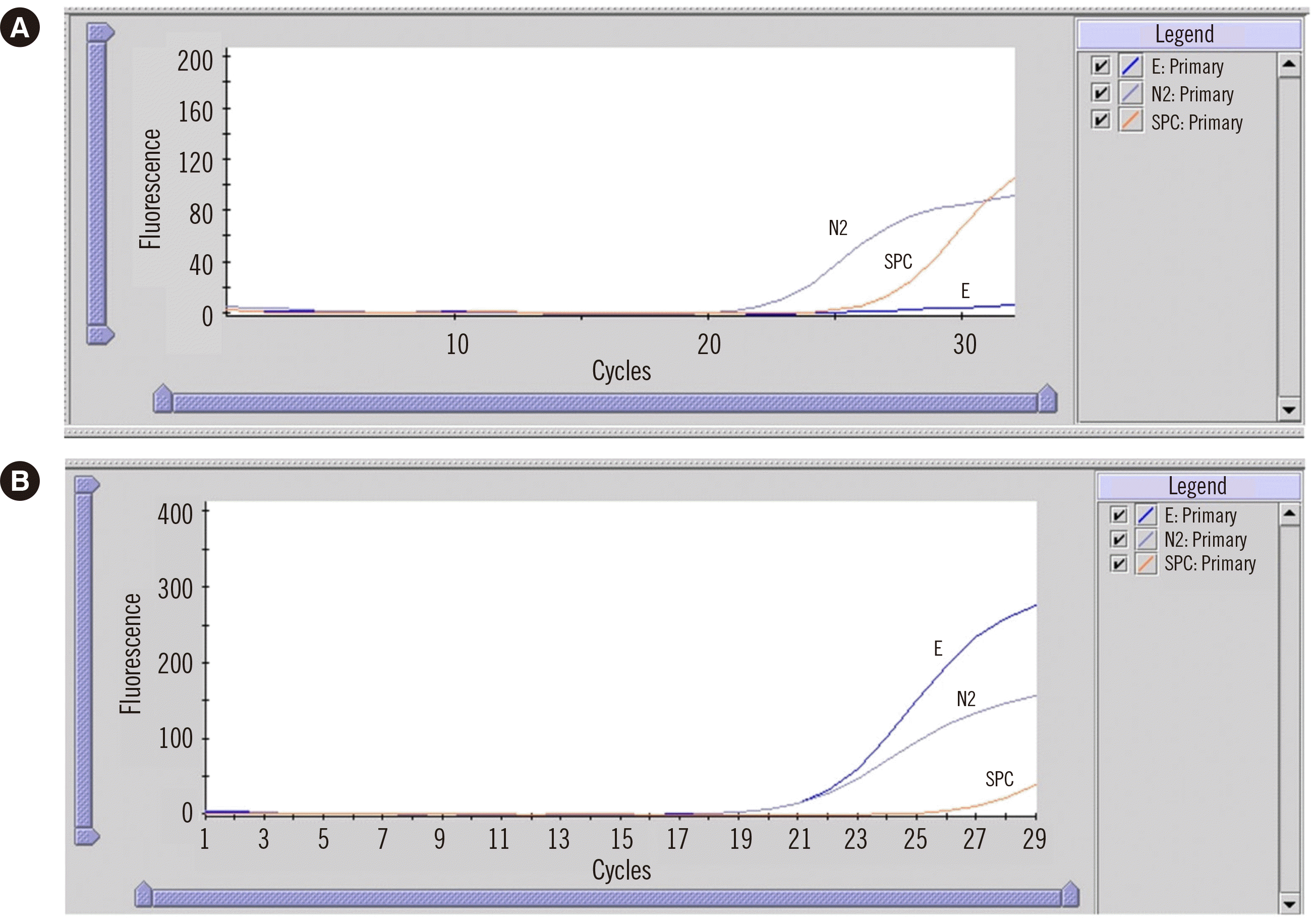
A 71-year-old man diagnosed as having COVID-19 four days prior was admitted to the emergency room of our hospital with hypovolemic shock in September 2022. The patient’s medical history included liver transplantation due to hepatocellular carcinoma 16 years prior to this episode. A rapid SARS-CoV-2 test using a nasopharyngeal swab (NPS) sample and the Xpert assay yielded inconclusive results, with positive N2 detection (Ct value of 25.5) but negative E gene detection (Fig. 1A). The sample was re-tested using the cobas SARS-CoV-2 and Influenza A/B Test (Roche Molecular Systems, Branchburg, NJ, USA), with Ct values of 22.82 and 22.78 for E and RdRp, respectively. The patient recovered and was discharged on the fourth day of hospitalization.
The remaining NPS sample was stored at –70°C, and nucleic acids were extracted using the eMAG platform (bioMérieux, Marcy-l’Étoile, France). After reverse transcription using the SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific, Vilnius, Lithuania), the E gene region was amplified and sequenced using the following primers (https://sars-cov2-sanger-primer-lookup.thermofisher.com/): SC2M1-67_LEFT (5´-AAAATTGTTGATGAGCCTGAAGAACA-3´) and SC2M1-67_RIGHT (5´-ACTAGGTTCCATTGTTCAAGGAGC-3´). The thermal cycles were as follows: polymerase activation at 95°C for 10 minutes followed by 40 cycles of 96°C for 3 seconds, 62°C for 15 seconds, and 68°C for 30 seconds. In comparison with the genome sequence of SARS-CoV-2 isolate Wuhan-Hu-1 (NC_045512.2), only one single-nucleotide polymorphism was found: NC_045512.2 (E_v004):c.26C>T (p.Thr9Ile). This variant is frequently reported in SARS-CoV-2 Omicron variants (https://www.uniprot.org/uniprotkb/P0DTC4/entry). However, no changes that could account for the gene dropout were observed in the E gene region. Therefore, the original sample was re-tested using the Xpert assay, this time yielding positive Ct values of 22.82 and 22.78 for E and N2, respectively (Fig. 1B).
We report a puzzling SARS-CoV-2 Xpert assay result, initially being positive for N2 but negative for E. This phenomenon, referred to as “gene dropout,” can be caused by genetic alterations as well as low sample quality and/or low viral load [3, 8]. The variant c.26C>T (p.Thr9Ile) is unlikely to cause false-negative results, considering its high frequency of 46.8% among Korean sequences when searched on CoVal (https://coval.ccpem.ac.uk/). In our case, the false-negative result may have been due to a technical issue—specifically, instability of the reagent in the instrument’s cartridge—as retesting of the original sample yielded positive detection of both targets (N and E). Moreover, the Xpert EV assay, from the same company, was recalled due to a cartridge problem in the summer of 2022, which adds to the suspicion regarding the instability of the reagent used in the Xpert assay. This case highlights the advantage of a multi-target approach for SARS-CoV-2 testing, even in conserved target regions. We did not confirm the genetic lineage of SARS-CoV-2 in this case, which is a limitation of our study.
In summary, not only novel genetic variants but also reagent instability can cause gene dropout. The adoption of multi- rather than single-target analysis in SARS-CoV-2 assays can reduce false-negatives and avoid COVID-19 misdiagnosis.
Notes
AUTHOR CONTRIBUTIONS
Conceptualization: Sung H. Data curation: Park K and Sung H. Funding acquisition: Sung H. Investigation: Park K. Supervision: Sung H and Kim MN. Writing—original draft: Park K. Writing—review & editing: Sung H and Kim MN. All authors reviewed and approved the final version of the manuscript.
REFERENCES
1. Kim YJ, Sung H, Ki CS, Hur M. 2020; COVID-19 testing in South Korea: current status and the need for faster diagnostics. Ann Lab Med. 40:349–50. DOI: 10.3343/alm.2020.40.5.349. PMID: 32237287. PMCID: PMC7169622.

2. Tahan S, Parikh BA, Droit L, Wallace MA, Burnham CD, Wang D. 2021; SARS-CoV-2 E gene variant alters analytical sensitivity characteristics of viral detection using a commercial reverse transcription-PCR assay. J Clin Microbiol. 59:e0007521. DOI: 10.1128/JCM.00075-21. PMID: 33903167. PMCID: PMC8218754.

3. Vanaerschot M, Mann SA, Webber JT, Kamm J, Bell SM, Bell J, et al. 2020; Identification of a polymorphism in the N gene of SARS-CoV-2 that adversely impacts detection by reverse transcription-PCR. J Clin Microbiol. 59:e02369–20. DOI: 10.1128/JCM.02369-20. PMID: 33067272. PMCID: PMC7771441.

4. Rhoads DD, Plunkett D, Nakitandwe J, Dempsey A, Tu ZJ, Procop GW, et al. 2021; Endemic SARS-CoV-2 polymorphisms can cause a higher diagnostic target failure rate than estimated by aggregate global sequencing data. J Clin Microbiol. 59:e0091321. DOI: 10.1128/JCM.00913-21. PMID: 33972454. PMCID: PMC8288267.

5. Ziegler K, Steininger P, Ziegler R, Steinmann J, Korn K, Ensser A. 2020; SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 25:2001650. DOI: 10.2807/1560-7917.ES.2020.25.39.2001650. PMID: 33006300. PMCID: PMC7531073.

6. Hong KH, In JW, Lee J, Kim SY, Lee KA, Kim S, et al. 2022; Prevalence of a single-nucleotide variant of SARS-CoV-2 in Korea and its impact on the diagnostic sensitivity of the Xpert Xpress SARS-CoV-2 assay. Ann Lab Med. 42:96–9. DOI: 10.3343/alm.2022.42.1.96. PMID: 34374354. PMCID: PMC8368233.

7. Schizas N, Michailidis T, Samiotis I, Patris V, Papakonstantinou K, Argiriou M, et al. 2020; Delayed diagnosis and treatment of a critically ill patient with infective endocarditis due to a false-positive molecular diagnostic test for SARS-CoV-2. Am J Case Rep. 21:e925931-1-3. DOI: 10.12659/AJCR.925931. PMID: 32980852. PMCID: PMC7526942.

8. Kanji JN, Zelyas N, MacDonald C, Pabbaraju K, Khan MN, Prasad A, et al. 2021; False negative rate of COVID-19 PCR testing: a discordant testing analysis. Virol J. 18:13. DOI: 10.1186/s12985-021-01489-0. PMID: 33422083. PMCID: PMC7794619.

Fig. 1
Amplification curves for SARS-CoV-2 generated by the reverse transcription-based Xpert Xpress assay (Cepheid, Sunnyvale, CA, USA). (A) Amplification curves for the sample tested upon admission of the patient to the emergency room and (B) for the same sample re-tested for confirmation.
Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download