The genus
Chlamydia currently comprises 11 species:
C. abortus,
C. avium,
C. caviae,
C. felis,
C. gallinacea,
C. muridarum,
C. pecorum,
C. pneumoniae,
C. psittaci,
C. suis, and
C. trachomatis [
1]. This diverse group of obligate intracellular gram-negative bacteria has a well-characterized biphasic lifecycle and infects nearly all phylogenetic groups of animals [
2-
4]. At least six species are capable of infecting humans and exhibit a broad range of infections, including urogenital and respiratory infections, spontaneous abortion, and conjunctivitis [
5].
C. pneumoniae and
C. psittaci are closely related respiratory pathogens that cause significant morbidity and mortality in humans [
6-
8]. The primary risk factor for infection and localized outbreaks is direct exposure to various bird species, which typically occurs in closed settings where birds are housed or handled.
The diagnosis of
Chlamydia infections is challenging because of the intracellular nature of the bacteria, requirement of cell culture for isolation, and close relatedness of the species comprising the genus [
9]. Serology is the most widely available assay method in commercial laboratories; however, sensitivity and specificity problems with these assays are well documented and may impact diagnosis [
10]. Culture is rarely performed because of the technical difficulty and safety concerns with handling infectious material [
6]. Strategies that rely on nucleic acid amplification, such as
ompA sequencing, restriction fragment length polymorphism analysis, and real-time PCR, have significantly advanced
Chlamydia diagnosis [
11]. Despite the widespread use of real-time PCR assays for detecting most other respiratory pathogens, including
C. pneumoniae, this method is not widely available in clinical laboratories for diagnosing
C. psittaci infections. The current diagnostic criteria for confirmed and probable cases rely solely on culture and serological methods, neither of which is reasonably suited for the diagnosis of acute infections [
6]. The development and widespread implementation of sensitive and specific real-time PCR assays for the detection of
C. psittaci in human clinical specimens would improve accuracy of diagnosing related infections.
The widely expanded use of next-generation sequencing in recent years has resulted in a steady increase in high-quality publicly available genome sequencing data. This data can be used to run and evaluate existing assays and identify new targets to characterize the
Chlamydia species with higher confidence. For example,
C. abortus genome sequences recently submitted to the National Center for Biotechnology Information genome database (GenBank) revealed that genetic sequences identified by Voigt,
et al. [
12], originally thought to be present only in
C. psittaci, are actually shared with several
C. abortus strains. The target sequence for detecting
C. psittaci in our previously published real-time PCR assay was discovered in one of these identified regions, rendering the assay inadequate for specific
C. psittaci detection [
11].
From September 2018 to October 2019, we reanalyzed all publicly available complete genomes to identify new target sequences specific to
C. psittaci and areas conserved among all
Chlamydia species. Approval from the Institutional Review Board was not required for this process since these genomes were publicly available. Through bioinformatics analysis, we identified a 128-bp sequence in a tyrosine tRNA gene (accession number CP002586.1, gene G5O_0561) that is present in all currently available
Chlamydia genomes (
Supplemental Data Fig. S1). We also identified a
C. psittaci-specific sequence in a gene encoding a polymorphic membrane protein (accession number CP002586.1, coding sequence AEB55237.1). Primers and 5´ hydrolysis (TaqMan) probes for target amplification were manually designed and synthesized (Thermo Fisher Scientific Waltham, MA, USA;
Supplemental Data Table S1). Primer and probe sets were initially evaluated and individually optimized (data not shown). A multiplex real-time PCR assay was designed and optimized for performance in comparison with established assays for the detection of
C. pneumoniae, using human RNaseP as a specimen quality and extraction control [
13,
14]. The multiplex real-time PCR mixture consisted of 5 µL of nucleic acid template, 12.5 µL of PerfeCTa Multiplex qPCR SuperMix (Quanta Biosciences, Gaithersburg, MD, USA), primers and probes at the working concentrations listed in
Supplemental Data Table S1, and nuclease-free water (Promega, Madison, WI, USA) to achieve a final volume of 25 µL. PCRs were run on the Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA), at the following cycling conditions: 95°C for 5 minutes followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute.
Genomic DNA extracted from purified
C. psittaci and
C. pneumoniae elementary bodies as previously described [
15] was adjusted to a concentration of 1 ng/µL, combined with 100 ng human nucleic acid (Promega), and amplified; representative real-time PCR amplification curves are shown in
Fig. 1. The specificity of the multiplex assay was verified using a panel of 10 characterized
C. psittaci isolates representing genotypes A, B, C, D, E, F, and G, eight
C. pneumoniae isolates, three
C. trachomatis isolates representing three genotypes, six other
Chlamydia species, and 22 other bacterial and viral pathogens commonly found in respiratory specimens (
Table 1). All results were interpreted based on the algorithm presented in
Supplemental Data Table S2.
Fig. 1
Amplification plots from the multiplex real-time PCR assay for the detection of all Chlamydia species (orange), Chlamydia psittaci (green), Chlamydia pneumoniae (blue), and human DNA (red). All specimens were tested in triplicate. All channels are shown to demonstrate the assay specificity and lack of spectral crossover.
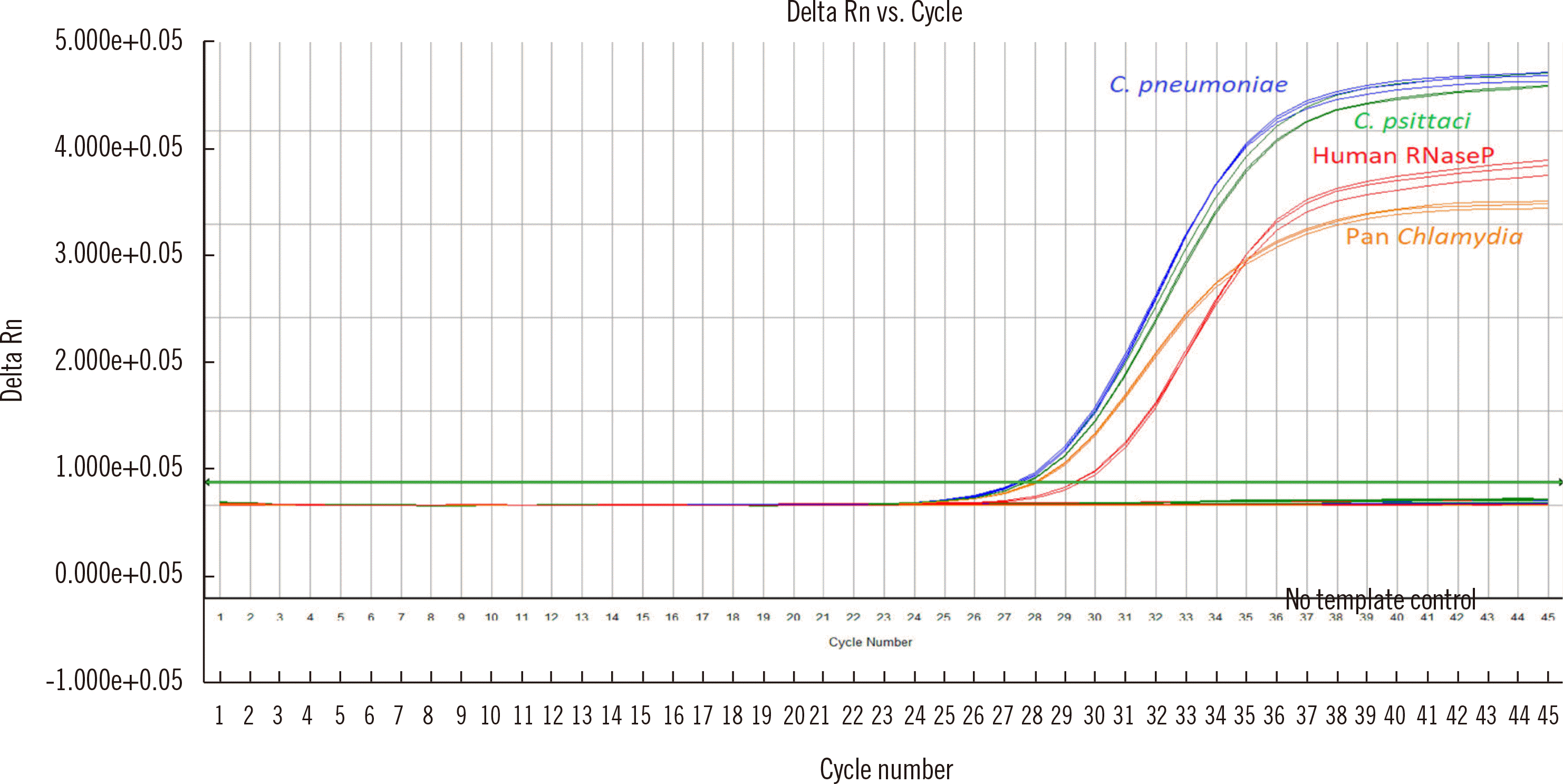

Table 1
Multiplex assay results for Chlamydia and non-Chlamydia isolates
|
Source |
Strain name/ATCC accession No. (species name) |
Pan-Chlamydia PCR result |
Chlamydia psittaci PCR result |
Chlamydia pneumoniae PCR result |
Human RNaseP result |
|
ATCC |
DD34 (C. psittaci genotype A) |
+ |
+ |
– |
– |
|
CDC |
UGA P6 (C. psittaci genotype A) |
+ |
+ |
– |
– |
|
CDC |
LSU P8 (C. psittaci genotype A) |
+ |
+ |
– |
– |
|
CDC |
CP3 (C. psittaci genotype B) |
+ |
+ |
– |
– |
|
CDC |
GR9 (C. psittaci genotype C) |
+ |
+ |
– |
– |
|
CDC |
NJ1 (C. psittaci genotype D) |
+ |
+ |
– |
– |
|
CDC |
TT3 (C. psittaci genotype D) |
+ |
+ |
– |
– |
|
CDC |
Frances (C. psittaci genotype E) |
+ |
+ |
– |
– |
|
CDC |
VS225 (C. psittaci genotype F) |
+ |
+ |
– |
– |
|
CDC |
5840-1 (C. psittaci genotype G) |
+ |
+ |
– |
– |
|
CDC |
A03 (C. pneumoniae) |
+ |
– |
+ |
– |
|
ATCC |
TW183 (C. pneumoniae) |
+ |
– |
+ |
– |
|
CDC |
U1271 (C. pneumoniae) |
+ |
– |
+ |
– |
|
CDC |
KOAL (C. pneumoniae) |
+ |
– |
+ |
– |
|
CDC |
CWL011 (C. pneumoniae) |
+ |
– |
+ |
– |
|
CDC |
12-07974 (C. pneumoniae) |
+ |
– |
+ |
– |
|
CDC |
FML-12 (C. pneumoniae) |
+ |
– |
+ |
– |
|
CDC |
CM1 (C. pneumoniae) |
+ |
– |
+ |
– |
|
ATCC |
VR-628 (C. pecorum) |
+ |
– |
– |
– |
|
ATCC |
5-45-15 (C. suis) |
+ |
– |
– |
– |
|
ATCC |
VR-120 (C. felis) |
+ |
– |
– |
– |
|
ATCC |
VR-123 (C. muridarum) |
+ |
– |
– |
– |
|
ATCC |
VR-656 (C. abortus) |
+ |
– |
– |
– |
|
CDC |
C. trachomatis genotype A |
+ |
– |
– |
– |
|
CDC |
C. trachomatis genotype D |
+ |
– |
– |
– |
|
CDC |
C. trachomatis genotype E |
+ |
– |
– |
– |
|
ATCC |
VR-813 (C. caviae) |
+ |
– |
– |
– |
|
CDC |
Adenovirus |
– |
– |
– |
– |
|
CDC |
Bordetella pertussis
|
– |
– |
– |
– |
|
CDC |
Escherichia coli
|
– |
– |
– |
– |
|
Promega |
G3041 (human DNA) |
– |
– |
– |
+ |
|
CDC |
Group A Streptococcus |
– |
– |
– |
– |
|
CDC |
Group B Streptococcus |
– |
– |
– |
– |
|
CDC |
Haemophilus influenzae
|
– |
– |
– |
– |
|
CDC |
Influenza A H1N1 |
– |
– |
– |
– |
|
CDC |
Influenza A H3N2 |
– |
– |
– |
– |
|
CDC |
Influenza B |
– |
– |
– |
– |
|
CDC |
Klebsiella pneumoniae
|
– |
– |
– |
– |
|
CDC |
Legionella pneumophila
|
– |
– |
– |
– |
|
CDC |
Moraxella catarrhalis
|
– |
– |
– |
– |
|
ATCC |
Mycoplasma pneumoniae M129
|
– |
– |
– |
– |
|
CDC |
Neisseria meningitidis
|
– |
– |
– |
– |
|
CDC |
Pseudomonas aeruginosa
|
– |
– |
– |
– |
|
CDC |
Respiratory syncytial virus |
– |
– |
– |
– |
|
CDC |
Staphylococcus aureus
|
– |
– |
– |
– |
|
CDC |
Staphylococcus epidermidis
|
– |
– |
– |
– |
|
CDC |
Streptococcus agalactiae
|
– |
– |
– |
– |
|
CDC |
Streptococcus pneumoniae
|
– |
– |
– |
– |
|
CDC |
Streptococcus pyogenes
|
– |
– |
– |
– |

Amplification was observed for all
Chlamydia isolates with the pan-
Chlamydia assay and for all
C. psittaci genotypes and
C. pneumoniae isolates with each species-specific assay (
Table 1). No amplification was observed when testing no-template controls (N=100) consisting of nuclease-free water (data not shown). Analytical sensitivity was verified by testing a 10-fold dilution series of purified DNA from
C. psittaci strain 6BC (genotype A) and
C. pneumoniae TW-183 ranging from 10 ag/µL to 1 ng/µL. Limits of detection (LODs) were calculated using probit analysis [
16]. The LODs for the pan-
Chlamydia, -
C. psittaci, -
C. pneumoniae, and RNaseP assays were 8.66 fg/µL, 7.64 fg/µL, 9.02 fg/µL, and 2.67 pg/µL respectively. The LODs of all assays were comparable in single- and multiplex formats (data not shown).
Nasopharyngeal (NP) and oropharyngeal (OP) swabs and cerebrospinal fluid, stool, and sputum specimens (N=34) submitted to the Centers for Disease Control and Prevention (CDC) Pneumonia Response and Surveillance Laboratory during domestic respiratory disease outbreak investigations were used to evaluate the new multiplex assay. Ten specimens previously identified as positive for
C. psittaci by real-time PCR [
11] and
ompA sequencing [
10], 12
C. pneumoniae-positive specimens [
14], and 12 specimens negative for both
C. psittaci and
C. pneumoniae were included. Total nucleic acid was extracted from all clinical specimens using the MagNA Pure Compact Nucleic Acid Purification System and Total Nucleic Acid Isolation kit I (both from Roche Applied Bioscience, Indianapolis, IN, USA) according to manufacturer’s instructions, and all specimens were tested in triplicate.
The multiplex PCR assay results matched with historical assay results for all specimens, except one.
C. psittaci was detected in one NP swab that previously tested negative for this species. This NP swab was collected from a patient from whom a sputum specimen collected at the same time tested positive for
C. psittaci; therefore, the detection in the NP swab most likely was a true positive identified owing to a slight increase in sensitivity of the new
C. psittaci assay. All specimens tested positive for the human control RNaseP (
Supplemental Data Table S3). In addition, 2018 and 2019
C. psittaci and
C. pneumoniae respiratory external quality assessment (EQA) panels from Quality Control for Molecular Diagnostics (QCMD, Glasgow, UK) were tested using the new assay; the results were 100% accurate for all panel specimens (data not shown). The
C. psittaci EQA panel includes specimens that contain
Chlamydia species other than
C. pneumoniae and
C. psittaci; the
Chlamydia species target was detected in these specimens, whereas the two species-specific targets were not detected.
To our knowledge, this is the first report of a multiplex real-time PCR assay for the detection of all Chlamydia species and simultaneous differentiation of C. psittaci and C. pneumoniae, along with an internal control, for use in human clinical specimens. The assay is substantially easier, faster, and more reliable than traditional diagnostic methods, such as culture and serology. The main limitations of this study were the low number of primary specimens available and the lack of a comprehensive panel of Chlamydia species to test. However, the EQA panels and robust bioinformatics analysis support the accuracy and reliability of the assay method.
The multiplex assay will be useful for detecting previously unrecognized Chlamydia infections and will allow researchers and clinicians to better understand the role of novel and rare Chlamydia species in human disease. The assay is easily adaptable to the veterinary and agricultural fields, where several Chlamydia species remain poorly understood and difficult to detect because of the lack of reliable standardized diagnostic assays. Unlike traditional diagnostics, this assay can be easily applied to a diverse array of specimen types, and the inclusion of a pan-Chlamydia assay will aid in the detection of rare and novel Chlamydia infections with unusual clinical presentations in both humans and animals. While PCR is currently not widely used for the diagnosis of C. psittaci infection, increased availability of laboratory-developed assays like the current one and accumulation of clinical validation data are likely to lead to the use of real-time PCR as a primary diagnostic method for C. psittaci in humans.

