1. Danese S, Fiocchi C. Ulcerative colitis. N Engl J Med. 2011; 365:1713–1725.

2. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017; 389:1756–1770.

3. Mizoguchi E, Low D, Ezaki Y, Okada T. Recent updates on the basic mechanisms and pathogenesis of inflammatory bowel diseases in experimental animal models. Intest Res. 2020; 18:151–167.

4. Ooi CJ, Hilmi I, Banerjee R, et al. Best practices on immunomodulators and biologic agents for ulcerative colitis and Crohn’s disease in Asia. Intest Res. 2019; 17:285–310.

5. Watanabe K. Clinical management for small bowel of Crohn’s disease in the treat-to-target era: now is the time to optimize treatment based on the dominant lesion. Intest Res. 2020; 18:347–354.

6. Toedter GP, Blank M, Lang Y, Chen D, Sandborn WJ, de Villiers WJ. Relationship of C-reactive protein with clinical response after therapy with ustekinumab in Crohn’s disease. Am J Gastroenterol. 2009; 104:2768–2773.

7. Sands BE. Biomarkers of inflammation in inflammatory bowel disease. Gastroenterology. 2015; 149:1275–1285.

8. Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009; 136:65–80.

9. Pittayanon R, Lau JT, Leontiadis GI, et al. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology. 2020; 158:930–946.

10. Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006; 55:205–211.

11. Zuo T, Ng SC. The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front Microbiol. 2018; 9:2247.

12. Yilmaz B, Juillerat P, Øyås O, et al. Microbial network disturbances in relapsing refractory Crohn’s disease. Nat Med. 2019; 25:323–336.

13. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019; 4:293–305.

14. Neumann C, Blume J, Roy U, et al. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host-microbiota homeostasis. Nat Immunol. 2019; 20:471–481.

15. Mahajan R, Midha V, Singh A, et al. Incidental benefits after fecal microbiota transplant for ulcerative colitis. Intest Res. 2020; 18:337–340.

16. Sehgal K, Khanna S. Gut microbiome and checkpoint inhibitor colitis. Intest Res. 2021; 19:360–364.

17. Bamba S, Inatomi O, Nishida A, et al. Relationship between the gut microbiota and bile acid composition in the ileal mucosa of Crohn’s disease. Intest Res. 2022; 20:370–380.

18. Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis: a randomized study. N Engl J Med. 1987; 317:1625–1629.

19. Best WR, Becktel JM, Singleton JW, Kern F Jr. Development of a Crohn’s disease activity index. National Cooperative Crohn’s Disease Study. Gastroenterology. 1976; 70:439–444.
20. Frank DN, Robertson CE, Hamm CM, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011; 17:179–184.

21. Papa E, Docktor M, Smillie C, et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One. 2012; 7:e39242.

22. Satokari R. Contentious host-microbiota relationship in inflammatory bowel disease: can foes become friends again? Scand J Gastroenterol. 2015; 50:34–42.

23. Vester-Andersen MK, Mirsepasi-Lauridsen HC, Prosberg MV, et al. Increased abundance of proteobacteria in aggressive Crohn’s disease seven years after diagnosis. Sci Rep. 2019; 9:13473.

24. Alam MT, Amos G, Murphy A, Murch S, Wellington E, Arasaradnam RP. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020; 12:1.

25. Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014; 15:382–392.

26. Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004; 127:412–421.

27. Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007; 2:119–129.

28. Kolho KL, Korpela K, Jaakkola T, et al. Fecal microbiota in pediatric inflammatory bowel disease and its relation to inflammation. Am J Gastroenterol. 2015; 110:921–930.

29. Pilarczyk-Zurek M, Chmielarczyk A, Gosiewski T, et al. Possible role of Escherichia coli in propagation and perpetuation of chronic inflammation in ulcerative colitis. BMC Gastroenterol. 2013; 13:61.

30. Mylonaki M, Rayment NB, Rampton DS, Hudspith BN, Brostoff J. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis. 2005; 11:481–487.

31. Zhang SL, Wang SN, Miao CY. Influence of microbiota on intestinal immune system in ulcerative colitis and its intervention. Front Immunol. 2017; 8:1674.

32. Olbjørn C, Cvancarova Småstuen M, Thiis-Evensen E, et al. Fecal microbiota profiles in treatment-naïve pediatric inflammatory bowel disease: associations with disease phenotype, treatment, and outcome. Clin Exp Gastroenterol. 2019; 12:37–49.
33. Prosberg M, Bendtsen F, Vind I, Petersen AM, Gluud LL. The association between the gut microbiota and the inflammatory bowel disease activity: a systematic review and meta-analysis. Scand J Gastroenterol. 2016; 51:1407–1415.

34. O’Mahony L, Feeney M, O’Halloran S, et al. Probiotic impact on microbial flora, inflammation and tumour development in IL-10 knockout mice. Aliment Pharmacol Ther. 2001; 15:1219–1225.

35. Peran L, Camuesco D, Comalada M, et al. Preventative effects of a probiotic, Lactobacillus salivarius ssp. salivarius, in the TNBS model of rat colitis. World J Gastroenterol. 2005; 11:5185–5192.
36. Feighery LM, Smith P, O’Mahony L, Fallon PG, Brayden DJ. Effects of Lactobacillus salivarius 433118 on intestinal inflammation, immunity status and in vitro colon function in two mouse models of inflammatory bowel disease. Dig Dis Sci. 2008; 53:2495–2506.

37. Bossuyt P, Verhaegen J, Van Assche G, Rutgeerts P, Vermeire S. Increasing incidence of Clostridium difficile-associated diarrhea in inflammatory bowel disease. J Crohns Colitis. 2009; 3:4–7.

38. Singh H, Nugent Z, Yu BN, Lix LM, Targownik LE, Bernstein CN. Higher incidence of Clostridium difficile infection among individuals with inflammatory bowel disease. Gastroenterology. 2017; 153:430–438.

39. Rodemann JF, Dubberke ER, Reske KA, Seo DH, Stone CD. Incidence of Clostridium difficile infection in inflammatory bowel disease. Clin Gastroenterol Hepatol. 2007; 5:339–344.

40. Ananthakrishnan AN, Binion DG. Impact of Clostridium difficile on inflammatory bowel disease. Expert Rev Gastroenterol Hepatol. 2010; 4:589–600.
41. Maharshak N, Barzilay I, Zinger H, Hod K, Dotan I. Clostridium difficile infection in hospitalized patients with inflammatory bowel disease: prevalence, risk factors, and prognosis. Medicine (Baltimore). 2018; 97:e9772.
42. Ben-Horin S, Margalit M, Bossuyt P, et al. Combination immunomodulator and antibiotic treatment in patients with inflammatory bowel disease and Clostridium difficile infection. Clin Gastroenterol Hepatol. 2009; 7:981–987.

43. Peeters M, Joossens S, Vermeire S, Vlietinck R, Bossuyt X, Rutgeerts P. Diagnostic value of anti-Saccharomyces cerevisiae and antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease. Am J Gastroenterol. 2001; 96:730–734.

44. Andersen K, Vogt C, Blondin D, et al. Multi-detector CT-colonography in inflammatory bowel disease: prospective analysis of CT-findings to high-resolution video colonoscopy. Eur J Radiol. 2006; 58:140–146.

45. Scanlan PD, Shanahan F, O’Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J Clin Microbiol. 2006; 44:3980–3988.

46. Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010; 139:1844–1854.

47. Seksik P, Rigottier-Gois L, Gramet G, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003; 52:237–242.

48. Wang W, Chen L, Zhou R, et al. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol. 2014; 52:398–406.

49. Song YL, Liu CX, McTeague M, Summanen P, Finegold SM. Clostridium bartlettii sp. nov., isolated from human faeces. Anaerobe. 2004; 10:179–184.

50. Rosario D, Benfeitas R, Bidkhori G, et al. Understanding the representative gut microbiota dysbiosis in metformin-treated type 2 diabetes patients using genome-scale metabolic modeling. Front Physiol. 2018; 9:775.

51. Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013; 504:451–455.

52. Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013; 504:446–450.

53. Choi SI, Son JH, Kim N, et al. Changes in cecal microbiota and short-chain fatty acid during lifespan of the rat. J Neurogastroenterol Motil. 2021; 27:134–146.

54. Beaud D, Tailliez P, Anba-Mondoloni J. Genetic characterization of the beta- glucuronidase enzyme from a human intestinal bacterium, Ruminococcus gnavus. Microbiology (Reading). 2005; 151(Pt 7):2323–2330.

55. Joossens M, Huys G, Cnockaert M, et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011; 60:631–637.

56. Tap J, Cools-Portier S, Pavan S, et al. Effects of the long-term storage of human fecal microbiota samples collected in RNAlater. Sci Rep. 2019; 9:601.

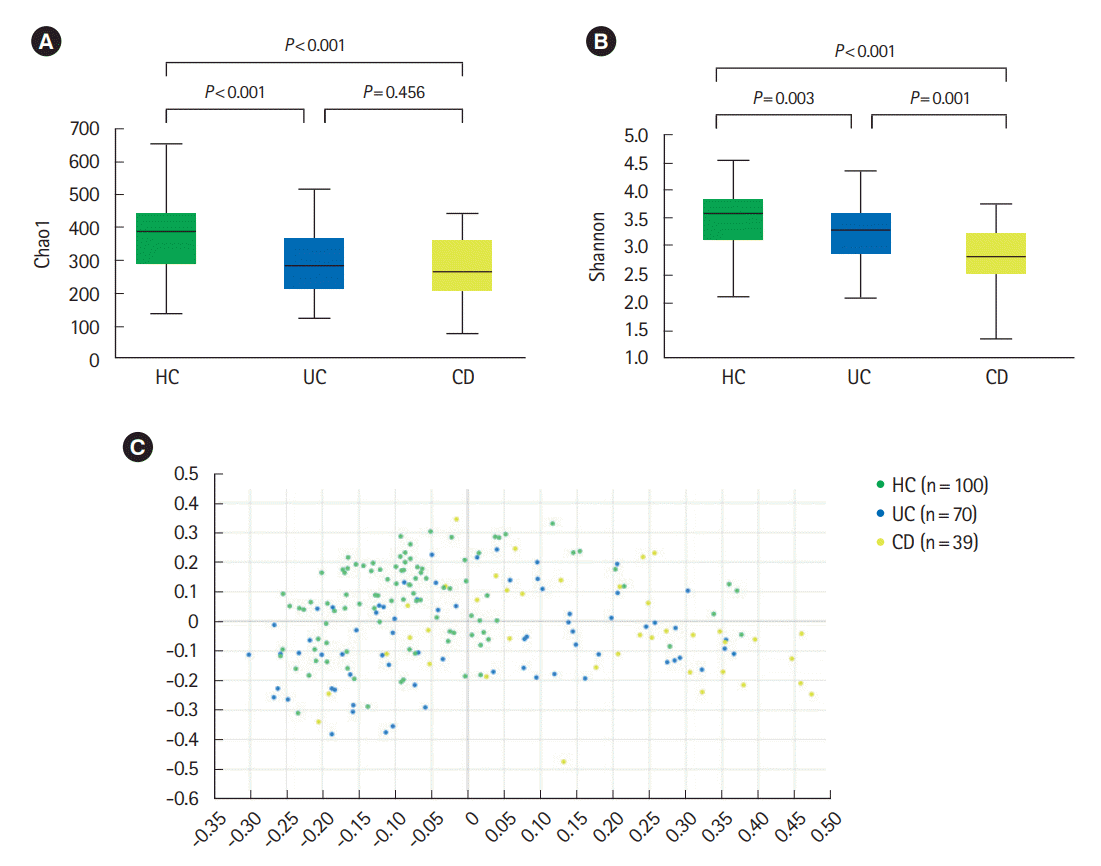
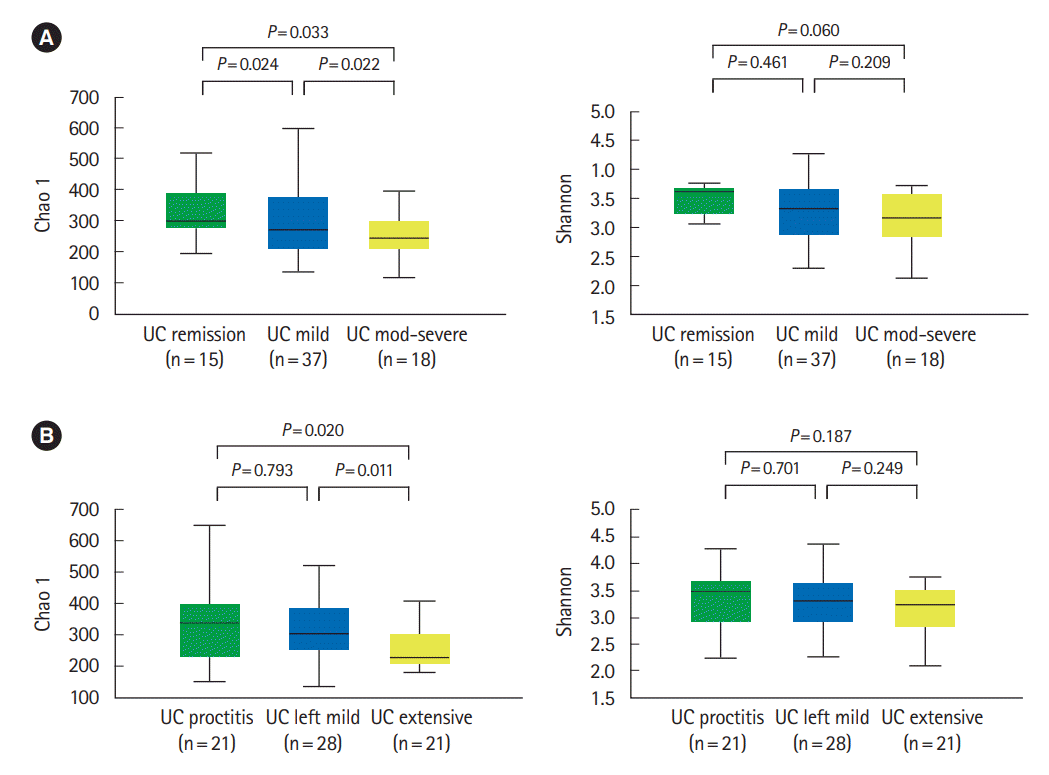
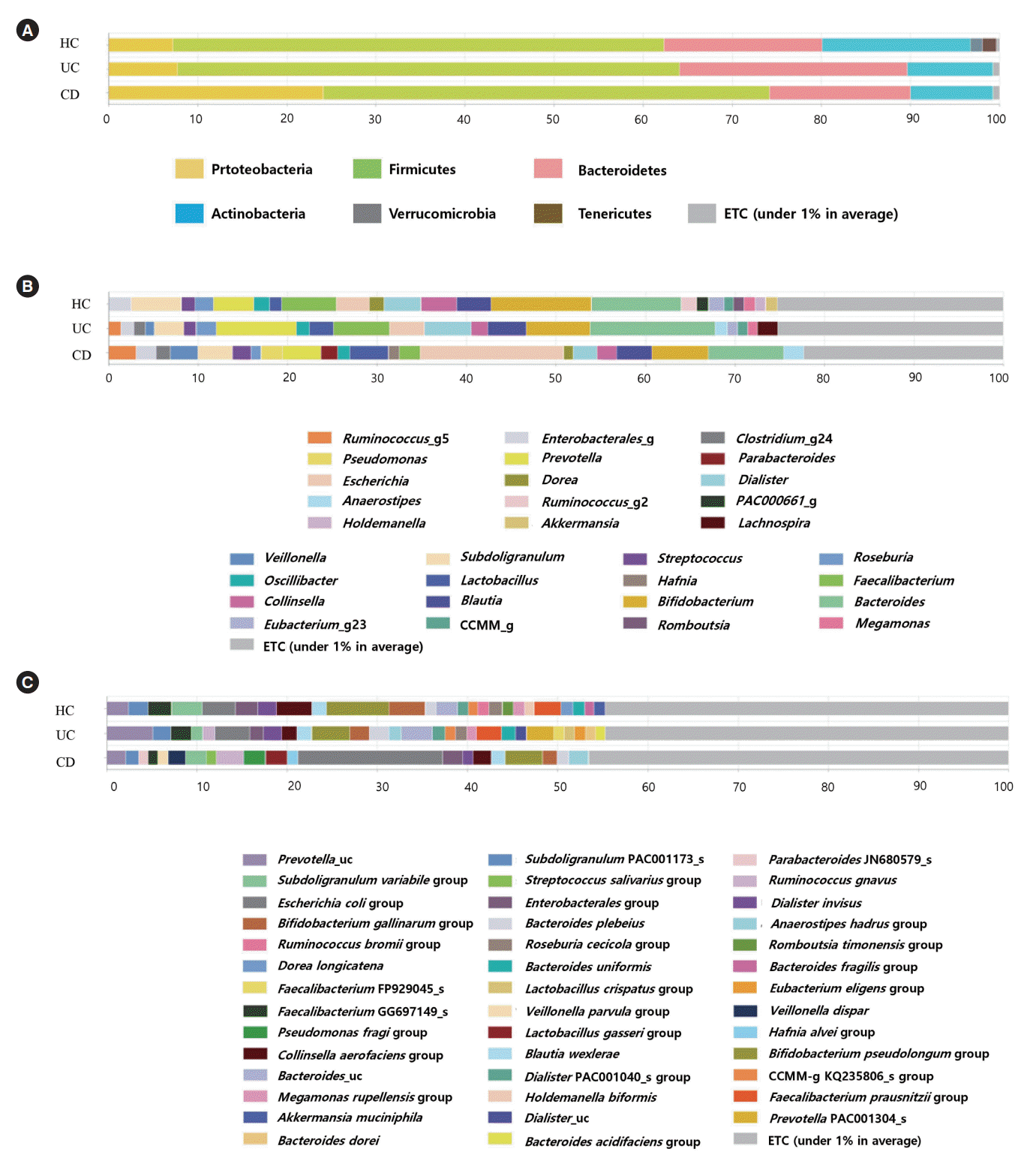
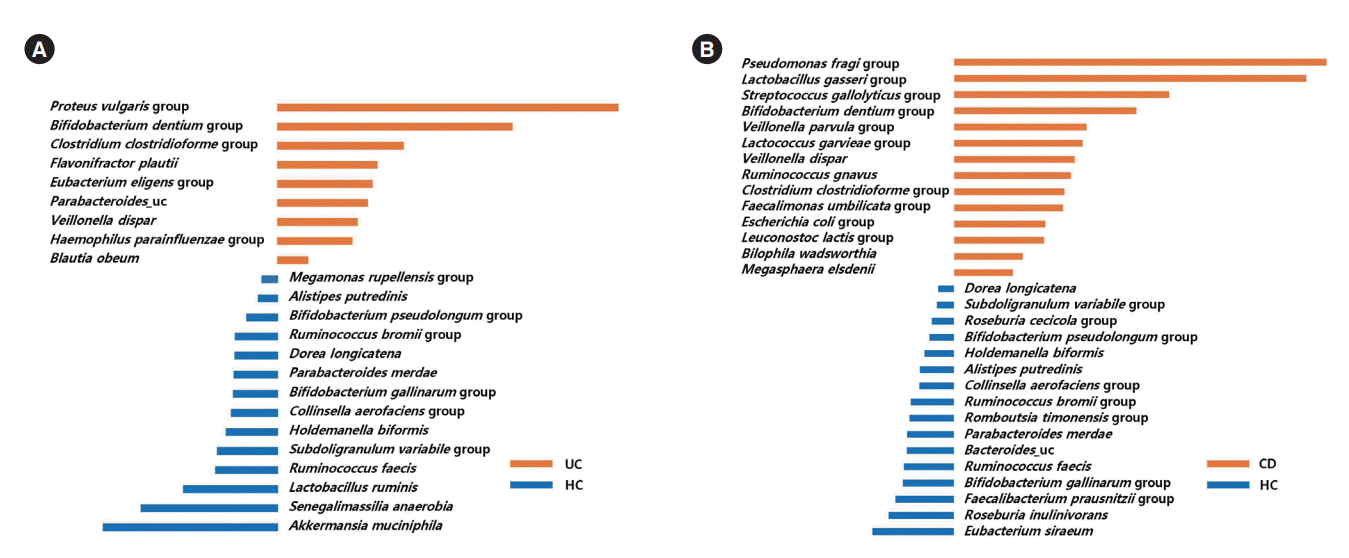




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download