Abstract
A role of gut microbiota in colorectal cancer (CRC) growth was first suggested in germ-free rats almost 50 years ago, and the existence of disease-associated bacteria (termed pathobionts) had becoming increasingly evident from experimental data of fecal transplantation, and microbial gavage or monoassociation. Altered bacterial compositions in fecal and mucosal specimens were observed in CRC patients compared to healthy subjects. Microbial fluctuations were found at various cancer stages; an increase of bacterial diversity was noted in the adenoma specimens, while a reduction of bacterial richness was documented in CRC samples. The bacterial species enriched in the human cancerous tissues included Escherichia coli, Fusobacterium nucleatum, and enterotoxigenic Bacteroides fragilis. The causal relationship of gut bacteria in tumorigenesis was established by introducing particular bacterial strains in in situ mouse CRC models. Detailed experimental protocols of bacterial gavage and the advantages and caveats of different experimental models are summarized in this review. The microbial genotoxins, enterotoxins, and virulence factors implicated in the mechanisms of bacteria-driven tumorigenesis are described. In conclusion, intestinal microbiota is involved in colon tumorigenesis. Bacteria-targeting intervention would be the next challenge for CRC.
An ecosystem of microorganisms habituates the human intestinal tract which is defined as the gut microbiota, composed of bacteria, archaea, virus, and fungi.1 The intestinal tract harbors around 1014 bacteria, with the highest amount in the colon.2
3
4 Over 1,000 bacterial species belonging mainly to 4 phyla were identified in humans, with each individual possessing more than 160 species.5 The collection of all genomes of the microbiota is termed microbiome, of which the number of intestinal microbial genes has been estimated to be ~100 times more than that of human genes.1
4
5
Accumulating evidence demonstrated that gut microbiota is involved in the maintenance of mucosal homeostasis, with an indispensable role in promotion of wound repair and epithelial barrier fortification.6
7
8
9 While a symbiotic relationship is established between commensal bacteria and the healthy host, the presence of an altered gut microbial population or the emergence of opportunistic bacteria from commensals may contribute to the development of IBD and colorectal cancer (CRC).10
11
12
13 Patients with IBD had higher risk of developing cancer later in life, termed colitis-associated CRC.14
15 It is generally believed the genetic and dietary factors, as well as chronic inflammation and altered microbial population predispose to tumor development.16
17
18
19
A relationship between commensal bacteria and intestinal carcinogenesis was first suggested in rodent models by Reddy et al. in 1974.20 Germ-free rats showed lower incidence of chemically induced duodenal and colonic tumors compared to those raised in conventional husbandry.20 Reduction of colonic tumor burden was also reported in several chemically induced or genetic deficient mouse models when derived under germ-free conditions.21
22
23 It should be kept in mind that the physiological relevance of germ-free models had been criticized since their mucosal immune systems were undeveloped due to the lack of microfloral establishment.24
25
26 Additional evidence of bacterial involvement in carcinogenesis was supported by studies showing reduction of tumor load after antibiotic manipulation of gut microbiota.22
27 The notion of disease-associated microbes originating from commensal-derived opportunistic bacteria (termed pathobionts) was shown by evidence of increased cancer growth in recipient mice after fecal transplantation from high tumor-bearing mouse.28
29 These studies suggested the existence of pathobionts with a protumoral characteristic in the fecal bacteria population.
On the other hand, presence of beneficial bacteria (termed probionts or probiotics) with tumor-suppressive metabolites was also found in experimental models.16
30 Short-chain fatty acids (SCFAs) such as butyrate are bacterial metabolites by fermentation of dietary fibers. Nutritional studies showed that high fiber diets or butyrate in the colon decreased the rate of aberrant crypt foci formation and reduced the tumor burden in animal models.31
32 A recent study using a gnotobiotic mouse model also demonstrated that supplementation with high dietary fiber and butyrate-producing bacteria significantly decreased colon tumor growth.30
Taken together, intestinal bacteria are involved in tumorigenesis with both detrimental and beneficial roles. Potential tumorigenic bacteria and their underlying mechanisms to promote cancer growth will be highlighted in this review. For more information on probiotics and cancer, please refer to other review articles.33
34
Despite similar fecal bacterial counts between healthy subjects and CRC patients,35 a clear difference in the composition of normal and tumor-associated microbiota was demonstrated. Numerous studies have shown reduction of microbial diversity in the stool samples and mucosal biopsies of patients with IBD36
37
38 (including CD and UC) and in those with CRC.39
40 In contrast, higher diversity and greater numbers of bacteria were noted in adenoma biopsies compared to healthy mucosa,41
42 suggesting that there existed dynamic changes of microbial population throughout the different tumor stages. It should be noted these cross-sectional studies are based on specimens collected at single time points. Therefore, the changes in gut microbiota only indicated a correlation but did not prove a cause-and-effect relationship for tumor formation in CRC patients.
The Intestinal microbes are mixed in the outer loose mucus layer and in chyme and feces. Many clinical studies have utilized stool samples for analysis of bacterial composition as a surrogate for intestinal microflora. Although the fecal bacteria may not represent the complete population of gut microbiota, this noninvasive approach has been widely practiced and conveyed crucial information of the bacterial community residing in the intestinal lumen. However, comparison of fecal microbiota composition to that of mucosal specimens and tumor biopsies may provide a more comprehensive view of the changes in the gut ecosystem.
Four main phyla were identified in the fecal microbiota of healthy individuals, including Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria, with Bacteroidetes and Firmicutes being the 2 predominant phyla that constitutes more than 90% of the total bacterial population.43
44 The percentages of Bacteroidetes and Firmicutes phyla in the fecal microbiota of healthy subjects were ~55% and ~40%, respectively.43
44 In CRC patients, these 2 phyla still constitutes the largest proportion in the stool microbiota.43
44 Being the 2 predominant phyla with higher total numbers in the individual, it is no doubt that alterations of the bacterial strains in the Bacteroidetes and Firmicutes phyla in the fecal population are easier to spot compared to other bacteria with lower counts. Nevertheless, accumulating evidence indicates that some bacteria belonging to the minor phyla in fecal microbiota may also play protumoral roles.
Under the Bacteroidetes phylum, enrichment of genera of Porphyromonas and Prevotella was reported in stool samples of CRC patients compared to healthy volunteers.44
45 Conflicting results were shown for the Bacteroides genus; increased percentage were reported in CRC patients in some studies,35
43 while others found a significant reduction.45 Among the Bacteroides species, a significant increase of Bacteroides fragilis and a decrease of Bacteroides vulgatus and Bacteroides uniformin were found in fecal samples of CRC patients.45
46
Under the Firmicutes phylum, increase of Enterococcus and Streptococcus genus and decrease of genera such as Faecalibacterium, Roseburia and Eubacterium and of Lachnospiraceae family were reported in stool samples of CRC patients compared to healthy volunteers.45
47 At the species levels, decreases in Ruminococcus obeum, Ruminococcus albus, Pseudobutyrivibrio ruminus, Lachnospira pectinoschiza, Lachnospira bovis were seen in the fecal microbiota of CRC patients.44 One of the common characteristics of the decreased bacterial genera and species in CRC patients belonging to the Firmicutes phylum is the ability of SCFA production.43
44
45 The altered stool bacterial profiling corresponded to the relatively lower levels of fecal butyrate in CRC patients compared to control subjects.44 In addition, no difference of Lactobacillus was shown in stool samples of cancer patients.35
45
46
48
Under the relatively minor phyla in fecal microbiota such as Proteobacteria, Actinobacteria and others, higher percentages of the genera of Escherichia (belonging to Proteobacteria phylum) and Fusobacterium (belonging to Fusobacteria phylum) were found in the stool samples of CRC patients compared to control subjects.43
45
46 The Bifidobacteria genus (belonging to Actinobacteria phylum) in fecal samples were either increased45 or showed no change35
46 in CRC patients.
Recent studies have utilized tumor/mucosal biopsies and lavage samples instead of fecal samples to validate and clarify the dysbiosis of gut microbiome. Data obtained from tumor biopsies indicated the alteration in mucosa-associated bacteria. Accumulating evidence showed distinct profiles between mucosal bacteria and fecal microbiota,47
49
50
51 whereas microfloral results obtained from mucosal biopsy and similar to those of lavage samples.52 For the evaluation of mucosal microbiota, tumor tissues are usually compared to normal mucosa of healthy individuals or to patient-matched non-cancerous tissues. One report had shown that Proteobacteria was the most predominant phylum (~60%) in the gut mucosa of healthy individuals.49 Another study showed the predominance of Firmicutes, Bacteroidetes, and Proteobacteria, of which the 3 phyla accounts for 80% of the microbiota in the control subjects.50 The findings showed that fecal microbiota as a surrogate only partially reflect the composition of the mucosa-associated bacteria.
By using mucosal specimens, the genera of Escherichia and Fusobacterium were found significantly increased in CRC patients compared to healthy controls.49
50
51 It is noteworthy that Escherichia and Fusobacterium constitute a minority of the healthy stool microbiome,43
44 and are also of low percentages (0.22% and 0.01%, respectively) of the bacterial population in healthy gut mucosa.49 However, a 15- and 1,000-fold increase of the percentage of Escherichia and Fusobacterium were noted in the microflora of biopsy tissues of CRC patients compared to that of healthy controls.49 When comparing patient-matched cancerous samples and adjacent non-cancerous tissues, abundance of Fusobacterium was also observed in tumors.49
50
51 The enrichment of Escherichia and Fusobacteria in tumor biopsies is in keeping with the findings in stool samples of CRC patients.43
45
46
Higher levels of Bacteroides genus was found in the mucosal microbiota of CRC patients than healthy controls.47 When comparing microbial composition in tumor biopsies to adjacent non-cancerous tissues, the percentage of Bacteroides were decreased in one report50 and increased in another study.49 The inconsistent result of Bacteroides in tumor tissues seems to reflect the fecal microbial composition which also shows conflicting data in the literatures.35
43
45
Decreased mucosal levels of Firmicutes and Actinobacteria phyla were observed in cancerous tissues of CRC patients compared to normal mucosa of healthy subjects or to adjacent non-cancerous tissues.49
51 A downregulation of Lachnospiraceae and Ruminococcaceae was observed in cancerous tissues compared to adjacent non-cancerous tissues, which also correlated with the finding of decreased bacterial genus in fecal samples of CRC patients.45
50 The data of mucosa-associated Lactobacillus and Bifidobacterium were inconsistent and a few reports showed no difference of these bacteria between tumor and control samples.35
48
Overall, the aforementioned studies all pointed to microbiota dysbiosis in fecal and mucosal samples of CRC patients despite inconsistency of the wax or wane of particular bacterial taxa. The fecal bacterial profiling only partially reflected mucosal microbiota in healthy controls and in CRC. Moreover, a higher ratio of the Proteobacteria and Fusobacteria phyla was observed in CRC patients when the analysis was based on mucosal specimens.
Clinical studies indicated a positive correlation between mucosal bacteria and disease pathogenesis. The gut commensals mostly reside in the intestinal lumen in physiological conditions, separated by the inner mucus layer and are rarely in direct contact with the epithelial cells.53
54
55
56 The concept of bacterial adhesion to host epithelial cells was first reported in uropathogenic Escherichia coli in 1908.57 In the past two decades, high numbers of mucosa-associated bacteria were reported in clinical studies of IBD and CRC. Enrichment of mucosa-associated E. coli was found in tissue biopsy of patients with CD and CRC.38
58
59
60
61
62 Abundance of mucosa-associated Fusobacterium was noted in CRC patients compared to healthy individuals.49
50 Invasive strains of Fusobacterium nucleatum were also isolated from biopsy specimens of CD or acute appendicitis.63
64
65 Moreover, higher levels of enterotoxigenic B. fragilis were found in colonic tumor samples obtained from patients.66
67 Evidence of the protumorigenic potentials of E. coli, F. nucleatum, and B. fragilis by using experimental models are discussed. The detailed experimental protocols of bacterial gavage in in situ mouse CRC models are listed in Table 1
58
68
69
70
71
72
73
74
75
76
77
78
79 and the advantages and caveats of different experimental models are summarized as followed.
Adherent-invasive E. coli (AIEC) was first isolated from ileal lesions in CD patients.80
81
82
83
84 Although AIEC was unable to colonize the intestine of wild type mice, the colitogenic activity of AIEC was shown in transgenic mice expressing human carcinoembryonic antigen-related cell adhesions molecules (CEACAMs; a receptor for type-1 pili or fimbriae [fim]).85 The data suggested that mucosal colonization of AIEC through fimbriae-mediated adhesion was a crucial step for its colitogenic ability as a pathobiont.85 However, there remains no direct evidence for the involvement of AIEC in colon cancer development, except the general link between inflammation and tumorigenesis. It is noteworthy that the fimbrial adhesin is historically known as a common feature for host colonization by various strains of gram-negative bacteria.57 Although fimbriae-mediated adhesion also affected bacterial uptake or promote proinflammatory response in epithelial cells,57 it has not been traditionally regarded as a strain-specific pathogenic virulence.86
87 Previous studies demonstrated increased tumor burden in human CEACAM-transgenic mice after azoxymethane (AOM) injections,88 but did not specify the bacterial strains responsible for the fimbriae-dependent tumor growth. It remains undetermined whether the adherent or invasive virulence of E. coli is involved in colorectal tumorigenesis.
A clear evidence of E. coli involvement in CRC development was reported with the use of 3 bacterial strains (i.e., NC101, CCR20, and 11G5) which harbors genotoxin-encoding polyketide synthase (pks) pathogenicity islands (Table 1).68
69
71
72 The colibactin produced by pks+
E. coli induced DNA damage and cellular senescence in intestinal epithelial cells.68
69
71
72 The pks+ NC101 strain was a mouse adherent-invasive E. coli, and the CCR20 and 11G5 strains were clinical isolates from tumor biopsies of CRC patients. Increased colonic tumor load was observed by monoassociation of E. coli NC101 in gnotobiotic interleukin 10 (IL-10)−/− mice given injections of AOM68
69 and in adenomatous polyposis coli (APC)Min/+-IL-10−/− mice.70 Moreover, higher tumor susceptibility was found after oral gavage of pks+
E. coli CCR20 to wild type mice administered AOM/dextran sulfate sodium (DSS).71
72 Increased tumor numbers and volume were documented in APCMin/+ mice following oral inoculation of pks+
E. coli 11G5 compared to the K12 strain.58 Details of the experimental design are described as followed and also summarized in Table 1. The germ-free IL-10−/− and APCMin/+-IL-10−/− mice were orally gavaged with 108 colony-forming units (CFU) of E. coli NC101 strain in one bolus for bacterial monoassociation.68
69
70 For oral gavage of E. coli CCR20 strain, the wild type mice were raised on specific pathogen free (SPF) conditions and given intraperitoneal injection of AOM, followed by pretreatment of streptomycin (a bactericidal antibiotic to G(−) bacteria to disturb microflora) in drinking water for 2 days prior to gavage of 109 CFU of E. coli CCR20 to facilitate bacterial colonization, and then subjected to 2 cycles of DSS in drinking water to induce colitis.71
72 In addition, APCMin/+ raised in SPF conditions were gavaged with 108 CFU of E. coli 11G5 strain 3 days after streptomycin treatment to facilitate bacterial colonization, and tumors were inspected at 7 weeks after infection.58 Due to the nature of these intestinal isolated E. coli which are not pathogens per se, microbiota disturbance by antibiotics prior to bacterial gavage or monoassociation in gnotobiotic mice are necessary to facilitate bacterial colonization to understand their effects on tumor development. Overall, higher tumor loads were found in the mouse groups given pks+
E. coli compared to the pks-deleted bacteria or pks-negative strains, indicating a crucial role of pks operon in promoting carcinogenesis.68
69
70
71
Other reports have demonstrated that the human CRC-associated E. coli (11G5 strain) induced colitis and increased crypt cell proliferation in transgenic mice with epithelial overexpression of human CEACAM.73 The E. coli 11G5 strain was gavaged at 2×108 CFU twice a week for 3 weeks after streptomycin and 0.25% DSS treatment in the human CEACAM-transgenic mice.73 Moreover, the human CRC-associated E. coli 11G5 triggered the production of cyclooxygenase (COX)-2 in macrophages after phagocytosis in vitro, in a pks-independent manner.74 Furthermore, human CRC-associated E. coli obtained from tumor biopsies were composed of genotoxin-positive and -negative populations.89 The pks+
E. coli were not adhesive, whereas the highly adherent E. coli were devoid of known genotoxins but caused DNA damage in vitro.89 The findings suggested that aside from causing genotoxicity, the induction of tumor-infiltrating macrophages and other unknown mechanisms may also play indispensable roles in E. coli-driven tumorigenesis.
Enrichment of F. nucleatum was demonstrated in the stool and tissue samples of CRC patients.43
49
50
66
90
91
92
F. nucleatum is known as a commensal G(−) species residing in the oral cavity, and has been reported as an invasive strain associated with inflammatory disease in the mouth such as gingivitis.93
94 Invasive strains of F. nucleatum were also isolated from inflamed biopsy tissues from patients with CD or acute appendicitis.63
64
65
Recent data showed that orogavage of F. nucleatum increased tumor burden in APCMin/+ mice or in wild type mice given AOM/DSS (Table 1).75
76
95 A number of mechanisms were proposed by in situ CRC mouse models, including β-catenin and nuclear factor-κB signaling via activation of Toll-like receptor 4 (TLR4) (a cell surface innate receptor for lipopolysaccharide [LPS]),75
95 and recruitment of tumor-infiltrating myeloid cells.76 Other studies by using human CRC cell lines and a xenograft model showed that FadA adhesin expressed on F. nucleatum, via extracellular binding to E-cadherin, induced nuclear translocation of β-catenin for oncogenic transcription and epithelial hyperproliferation.96
97 The invasiveness of F. nucleatum was not a requirement for its effect on oncogenic transcription but was associated with proinflammatory signals which could be indirectly involved in cancer development.96 So far, whether F. nucleatum infection and its pro-tumorigenic ability are associated with inflammation is still controversial. Although some reported that the histopathological features of colitis and enteritis were not induced,76 others showed upregulation of proinflammatory cytokines and chemokines following inoculation of F. nucleatum.75 It is noteworthy that the in situ CRC model was pre-fed with streptomycin (a bactericidal antibiotic to G(−) bacteria) to disturb the normal flora, followed by inoculation with a very high number of F. nucleatum for a long period of time (109 CFU by gavage per day for 20 weeks or 108 CFU by gavage per day for 8 weeks).75
76 With such a high bacterial load given continuously, the roles of F. nucleatum as a driver or a passenger for tumorigenesis remain in doubt.
A recent study demonstrated that gavage of a single strain or multiple strains of F. nucleatum obtained from clinical specimens of CRC patients to gnotobiotic APCMin/+ mice or APCMin/+-IL-10−/− mice did not increase colitis severity nor enhance colon tumor burden.70 The germ-free mice were gavaged weekly with a mixture of 6 F. nucleatum clinical isolates at 108 CFU per strain per mice, or were transferred to SPF microbiota prior to gavage of F. nucleatum mixtures and sacrificed 16 weeks later to examine tumor growth. However, no increase in tumors was observed in either protocols.70 The inconsistent data of F. nucleatum suggested that other unidentified mechanisms, such as interaction with other bacteria, may partly contribute to its protumoral characteristics.
A subclass of the human commensal Bacteroides species, enterotoxigenic Bacteroides fragilis (ETBF), was associated with acute inflammatory diarrheal disease and CRC in patients.66
98
99 Increased percentage of B. fragilis was reported in fecal microbiota.45
46 The levels of ETBF and enterotoxin gene (bft) in tumor and stool samples were significantly higher in late stages (III/IV) of CRC compared to control tissues.66
67
99 Despite reported as an obligate anaerobe, abundant literature showed invasive traits of the G(−) bacteria B. fragilis. The most frequent anaerobe isolated in clinical cases of peritonitis, intra-abdominal abscess or bacteremia is B. fragilis, suggesting its aerotolerance and invasiveness.100
101
102
Previous studies have demonstrated that ETBF triggered colitis and accelerated tumor growth in APCMin/+ mice (Table 1).77
78 The colonization of ETBF (108 CFU by oral gavage in one dose) was facilitated by pretreatment of clindamycin (an antibiotic against G(+) bacteria) and streptomycin (an antibiotic against G(−) bacteria) or gentamicin (a broad-spectrum antibiotic) to disturb the gut microbiota.77
78
79 Following antibiotic manipulation of microbiota, colonization with ETBF but not its nontoxigenic counterparts increased tumor burden in APCMin/+ mice.77
78 The B. fragilis enterotoxin (also known as fragilysin) acts as a metalloprotease that causes oxidative DNA damage, E-cadherin cleavage, epithelial barrier damage, and activation of STAT3/Th17 immune responses, and generation of protumoral monocytic myeloid suppressor cells.77
78
79
103
104 Studies in vitro showed that fragilysin stimulated the production of spermine oxidase in intestinal epithelial cell lines, suggesting a direct role of the enterotoxin on epithelial free radical production and DNA damage.78 Taken together, the findings indicated that ETBF was a strong colitogenic infectious agent that promoted tumorigenesis through both direct and indirect mechanisms.
A number of hints and questions arose through the review of protumorigenic pathobionts. The protumorigenic bacteria (e.g., E. coli, F. nucleatum, and B. fragilis) identified so far in in situ CRC models were all G(−) bacteria with mucosal colonization characteristics. First, the tumor-promoting bacteria are enriched in the mucosa and belong to the category of G(−) microbes, indicating that the juxtaposition of G(−) bacteria and its wall component (i.e., LPS) recognized by mucosal innate receptors are likely to contribute partly to the tumor susceptibility.33
105
106
107 The findings re-emphasized the role of abnormal expression or upregulation of epithelial immune receptors in tumor predisposition.107
108
109 Second, microbial colonization and active invasion was shown to facilitate other commensal bacteria to be internalized into epithelium,110
111
112 and may cause secondary bystanders to pass the epithelial barrier through paracellular spaces.113
114
115
116 The active or passive entry of bacteria could further fuel inflammation-associated tumor growth or may induce DNA damage and hyperproliferative signals in host cells. In addition, the common features of mucosal colonization among protumorigenic bacteria suggest that there may be shared adherence/invasion-associated virulence factors to induce epithelial damages, which may partly underlie bacterial mechanisms of epithelial malignant transformation. Third, physiological mucosal defense against pathogen colonization and invasion included free radical production117 and autophagy of infected organelles in the epithelial cells.118
119 The mechanisms underlying the incompentent clearance of intracellular microbes remain poorly understood. A recent paper suggested that increased levels of microRNA 106B caused a reduction of autophagy-related gene transcription in intestinal epithelia of CD may account for the failure of intracellular bacterial clearance.120 It remains unclear whether epithelial recognition of pathobionts derived from commensals may activate suppressive/tolerant or defensive mechanisms. The involvement of epithelial barrier impairment in tumorigenesis warrants further studies.
In summary, it is widely recognized nowadays that intestinal microbiota is involved in colorectal tumorigenesis. Protumorigenic bacteria (e.g., E. coli and F. nucleatum) which are not predominant species in fecal microflora are enriched in the cancerous tissues, and may promote tumorigenesis by expression of genotoxins and virulence factors. Further understanding of the host-microbe interaction and how the bacterial factors fit into the genetic and molecular paradigm of tumor development will shed light to the development of novel microbe-targeting therapy. Specific elimination of pathobionts with sparing of beneficial symbionts would be the next challenge.
Notes
References
1. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016; 375:2369–2379. PMID: 27974040.

2. Yu LC, Wang JT, Wei SC, Ni YH. Host-microbial interactions and regulation of intestinal epithelial barrier function: from physiology to pathology. World J Gastrointest Pathophysiol. 2012; 3:27–43. PMID: 22368784.

3. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006; 124:837–848. PMID: 16497592.

4. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016; 14:e1002533. DOI: 10.1371/journal.pbio.1002533. PMID: 27541692.

5. Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010; 464:59–65. PMID: 20203603.
6. Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017; 15:630–638. PMID: 28626231.

7. Owyang C, Wu GD. The gut microbiome in health and disease. Gastroenterology. 2014; 146:1433–1436. PMID: 24675436.

8. Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017; 17:271–285. PMID: 28303904.

9. Nagao-Kitamoto H, Kitamoto S, Kuffa P, Kamada N. Pathogenic role of the gut microbiota in gastrointestinal diseases. Intest Res. 2016; 14:127–138. PMID: 27175113.

10. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013; 13:800–812. PMID: 24132111.

11. Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap Adv Gastroenterol. 2013; 6:295–308.

12. Drewes JL, Housseau F, Sears CL. Sporadic colorectal cancer: microbial contributors to disease prevention, development and therapy. Br J Cancer. 2016; 115:273–280. PMID: 27380134.

13. Abreu MT, Peek RM Jr. Gastrointestinal malignancy and the microbiome. Gastroenterology. 2014; 146:1534–1546.e3. PMID: 24406471.

14. Munkholm P. Review article: the incidence and prevalence of colorectal cancer in inflammatory bowel disease. Aliment Pharmacol Ther. 2003; 18(Suppl 2):1–5.

15. Brackmann S, Andersen SN, Aamodt G, et al. Relationship between clinical parameters and the colitis-colorectal cancer interval in a cohort of patients with colorectal cancer in inflammatory bowel disease. Scand J Gastroenterol. 2009; 44:46–55. PMID: 18609187.

16. Bultman SJ. Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer. Mol Nutr Food Res. 2017; 61:1500902. DOI: 10.1002/mnfr.201500902.

17. Tözün N, Vardareli E. Gut microbiome and gastrointestinal cancer: les liaisons dangereuses. J Clin Gastroenterol. 2016; 50(Suppl):S191–S196. PMID: 27741173.
18. Hullar MA, Burnett-Hartman AN, Lampe JW. Gut microbes, diet, and cancer. Cancer Treat Res. 2014; 159:377–399. PMID: 24114492.

19. Hiley CT, Swanton C. Pruning cancer's evolutionary tree with lesion-directed therapy. Cancer Discov. 2016; 6:122–124. PMID: 26851181.

20. Reddy BS, Weisburger JH, Narisawa T, Wynder EL. Colon carcinogenesis in germ-free rats with 1,2-dimethylhydrazine and N-methyl-n'-nitro-N-nitrosoguanidine. Cancer Res. 1974; 34:2368–2372. PMID: 4843537.
21. Uronis JM, Mühlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitisassociated colorectal cancer susceptibility. PLoS One. 2009; 4:e6026. DOI: 10.1371/journal.pone.0006026. PMID: 19551144.

22. Zackular JP, Baxter NT, Iverson KD, et al. The gut microbiome modulates colon tumorigenesis. MBio. 2013; 4:e00692–e00613. DOI: 10.1128/mBio.00692-13. PMID: 24194538.

23. Li Y, Kundu P, Seow SW, et al. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APCMin/+ mice. Carcinogenesis. 2012; 33:1231–1238. PMID: 22461519.

24. Chung H, Pamp SJ, Hill JA, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012; 149:1578–1593. PMID: 22726443.

25. Vannucci L, Stepankova R, Kozakova H, Fiserova A, Rossmann P, Tlaskalova-Hogenova H. Colorectal carcinogenesis in germfree and conventionally reared rats: different intestinal environments affect the systemic immunity. Int J Oncol. 2008; 32:609–617. PMID: 18292938.

26. Rhee KJ, Sethupathi P, Driks A, Lanning DK, Knight KL. Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J Immunol. 2004; 172:1118–1124. PMID: 14707086.

27. Zackular JP, Baxter NT, Chen GY, Schloss PD. Manipulation of the gut microbiota reveals role in colon tumorigenesis. mSphere. 2015; 1:e00001–e00015. DOI: 10.1128/mSphere.00001-15. PMID: 27303681.

28. Couturier-Maillard A, Secher T, Rehman A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013; 123:700–711. PMID: 23281400.

29. Hu B, Elinav E, Huber S, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci U S A. 2013; 110:9862–9867. PMID: 23696660.

30. Donohoe DR, Holley D, Collins LB, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014; 4:1387–1397. PMID: 25266735.

31. Perrin P, Pierre F, Patry Y, et al. Only fibres promoting a stable butyrate producing colonic ecosystem decrease the rate of aberrant crypt foci in rats. Gut. 2001; 48:53–61. PMID: 11115823.

32. Singh N, Gurav A, Sivaprakasam S, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014; 40:128–139. PMID: 24412617.

33. So SS, Wan ML, El-Nezami H. Probiotics-mediated suppression of cancer. Curr Opin Oncol. 2017; 29:62–72. PMID: 27792053.

34. Zhu Y, Michelle Luo T, Jobin C, Young HA. Gut microbiota and probiotics in colon tumorigenesis. Cancer Lett. 2011; 309:119–127. PMID: 21741763.

35. Sobhani I, Tap J, Roudot-Thoraval F, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011; 6:e16393. DOI: 10.1371/journal.pone.0016393. PMID: 21297998.

36. Walker AW, Sanderson JD, Churcher C, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011; 11:7. PMID: 21219646.

37. Hansen R, Russell RK, Reiff C, et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am J Gastroenterol. 2012; 107:1913–1922. PMID: 23044767.

38. Lepage P, Häsler R, Spehlmann ME, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011; 141:227–236. PMID: 21621540.

39. Ahn J, Sinha R, Pei Z, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013; 105:1907–1911. PMID: 24316595.

40. Huipeng W, Lifeng G, Chuang G, Jiaying Z, Yuankun C. The differences in colonic mucosal microbiota between normal individual and colon cancer patients by polymerase chain reaction-denaturing gradient gel electrophoresis. J Clin Gastroenterol. 2014; 48:138–144. PMID: 24162169.

41. Shen XJ, Rawls JF, Randall T, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010; 1:138–147. PMID: 20740058.

42. Sanapareddy N, Legge RM, Jovov B, et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012; 6:1858–1868. PMID: 22622349.

43. Wu N, Yang X, Zhang R, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013; 66:462–470. PMID: 23733170.

44. Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013; 8:e70803. DOI: 10.1371/journal.pone.0070803. PMID: 23940645.

45. Wang T, Cai G, Qiu Y, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012; 6:320–329. PMID: 21850056.

46. Kasai C, Sugimoto K, Moritani I, et al. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol Rep. 2016; 35:325–333. PMID: 26549775.

47. Flemer B, Lynch DB, Brown JM, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017; 66:633–643. PMID: 26992426.

48. Gueimonde M, Ouwehand A, Huhtinen H, Salminen E, Salminen S. Qualitative and quantitative analyses of the Bifidobacterial microbiota in the colonic mucosa of patients with colorectal cancer, diverticulitis and inflammatory bowel disease. World J Gastroenterol. 2007; 13:3985–3989. PMID: 17663515.

49. Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. 2015; 6:20. PMID: 25699023.

50. Burns MB, Lynch J, Starr TK, Knights D, Blekhman R. Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med. 2015; 7:55. PMID: 26170900.

51. Gao R, Kong C, Huang L, et al. Mucosa-associated microbiota signature in colorectal cancer. Eur J Clin Microbiol Infect Dis. 2017; 36:2073–2083. PMID: 28600626.

52. Watt E, Gemmell MR, Berry S, et al. Extending colonic mucosal microbiome analysis-assessment of colonic lavage as a proxy for endoscopic colonic biopsies. Microbiome. 2016; 4:61. PMID: 27884202.

53. Yu LC. Commensal bacterial internalization by epithelial cells: an alternative portal for gut leakiness. Tissue Barriers. 2015; 3:e1008895. DOI: 10.1080/21688370.2015.1008895. PMID: 26451337.

54. Wu LL, Peng WH, Kuo WT, et al. Commensal bacterial endocytosis in epithelial cells is dependent on myosin light chain kinase-activated brush border fanning by interferon-gamma. Am J Pathol. 2014; 184:2260–2274. PMID: 24911373.

55. Johansson ME, Sjövall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol. 2013; 10:352–361. PMID: 23478383.

56. Johansson ME, Gustafsson JK, Holmén-Larsson J, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014; 63:281–291. PMID: 23426893.

57. Kline KA, Fälker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009; 5:580–592. PMID: 19527885.

58. Bonnet M, Buc E, Sauvanet P, et al. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res. 2014; 20:859–867. PMID: 24334760.
59. Prorok-Hamon M, Friswell MK, Alswied A, et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut. 2014; 63:761–770. PMID: 23846483.

60. Chassaing B, Rolhion N, de Vallée A, et al. Crohn disease: associated adherent-invasive E. coli bacteria target mouse and human Peyer's patches via long polar fimbriae. J Clin Invest. 2011; 121:966–975. PMID: 21339647.

61. Martin HM, Campbell BJ, Hart CA, et al. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004; 127:80–93. PMID: 15236175.

62. Kleessen B, Kroesen AJ, Buhr HJ, Blaut M. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol. 2002; 37:1034–1041. PMID: 12374228.

63. Strauss J, Kaplan GG, Beck PL, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011; 17:1971–1978. PMID: 21830275.

64. Swidsinski A, Dörffel Y, Loening-Baucke V, et al. Mucosal invasion by fusobacteria is a common feature of acute appendicitis in Germany, Russia, and China. Saudi J Gastroenterol. 2012; 18:55–58. PMID: 22249094.
65. Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun. 2011; 79:2597–2607. PMID: 21536792.

66. Viljoen KS, Dakshinamurthy A, Goldberg P, Blackburn JM. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS One. 2015; 10:e0119462. DOI: 10.1371/journal.pone.0119462. PMID: 25751261.

67. Boleij A, Hechenbleikner EM, Goodwin AC, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015; 60:208–215. PMID: 25305284.
68. Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012; 338:120–123. PMID: 22903521.

69. Arthur JC, Gharaibeh RZ, Mühlbauer M, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun. 2014; 5:4724. PMID: 25182170.

70. Tomkovich S, Yang Y, Winglee K, et al. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res. 2017; 77:2620–2632. PMID: 28416491.
71. Cougnoux A, Dalmasso G, Martinez R, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014; 63:1932–1942. PMID: 24658599.

72. Dalmasso G, Cougnoux A, Delmas J, Darfeuille-Michaud A, Bonnet R. The bacterial genotoxin colibactin promotes colon tumor growth by modifying the tumor microenvironment. Gut Microbes. 2014; 5:675–680. PMID: 25483338.

73. Raisch J, Buc E, Bonnet M, et al. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J Gastroenterol. 2014; 20:6560–6572. PMID: 24914378.
74. Raisch J, Rolhion N, Dubois A, Darfeuille-Michaud A, Bringer MA. Intracellular colon cancer-associated Escherichia coli promote protumoral activities of human macrophages by inducing sustained COX-2 expression. Lab Invest. 2015; 95:296–307. PMID: 25545478.
75. Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of microRNA-21. Gastroenterology. 2017; 152:851–866.e24. PMID: 27876571.
76. Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013; 14:207–215. PMID: 23954159.
77. Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009; 15:1016–1022. PMID: 19701202.
78. Goodwin AC, Destefano Shields CE, Wu S, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A. 2011; 108:15354–15359. PMID: 21876161.
79. Thiele Orberg E, Fan H, Tam AJ, et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2017; 10:421–433. PMID: 27301879.
80. Darfeuille-Michaud A, Neut C, Barnich N, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology. 1998; 115:1405–1413. PMID: 9834268.

81. Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004; 127:412–421. PMID: 15300573.

82. Mimouna S, Gonçalvès D, Barnich N, Darfeuille-Michaud A, Hofman P, Vouret-Craviari V. Crohn disease-associated Escherichia coli promote gastrointestinal inflammatory disorders by activation of HIF-dependent responses. Gut Microbes. 2011; 2:335–346. PMID: 22157238.
83. Martinez-Medina M, Aldeguer X, Lopez-Siles M, et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm Bowel Dis. 2009; 15:872–882. PMID: 19235912.

84. Baumgart M, Dogan B, Rishniw M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 2007; 1:403–418. PMID: 18043660.

85. Carvalho FA, Barnich N, Sivignon A, et al. Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J Exp Med. 2009; 206:2179–2189. PMID: 19737864.

86. Thanassi DG, Bliska JB, Christie PJ. Surface organelles assembled by secretion systems of Gram-negative bacteria: diversity in structure and function. FEMS Microbiol Rev. 2012; 36:1046–1082. PMID: 22545799.

87. Lillington J, Geibel S, Waksman G. Biogenesis and adhesion of type 1 and P pili. Biochim Biophys Acta. 2014; 1840:2783–2793. PMID: 24797039.

88. Chan CH, Cook D, Stanners CP. Increased colon tumor susceptibility in azoxymethane treated CEABAC transgenic mice. Carcinogenesis. 2006; 27:1909–1916. PMID: 16632476.

89. Buc E, Dubois D, Sauvanet P, et al. High prevalence of mucosaassociated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One. 2013; 8:e56964. DOI: 10.1371/journal.pone.0056964. PMID: 23457644.

90. Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012; 22:292–298. PMID: 22009990.

91. Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017; 66:70–78. PMID: 26408641.

92. Purcell RV, Pearson J, Aitchison A, Dixon L, Frizelle FA, Keenan JI. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One. 2017; 12:e0171602. DOI: 10.1371/journal.pone.0171602. PMID: 28151975.

93. Fredricks DN, Schubert MM, Myerson D. Molecular identification of an invasive gingival bacterial community. Clin Infect Dis. 2005; 41:e1–e4. DOI: 10.1086/430824. PMID: 15937752.

94. Matsuo T, Shirakami T, Ozaki K, Nakanishi T, Yumoto H, Ebisu S. An immunohistological study of the localization of bacteria invading root pulpal walls of teeth with periapical lesions. J Endod. 2003; 29:194–200. PMID: 12669880.

95. Chen Y, Peng Y, Yu J, et al. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget. 2017; 8:31802–31814. PMID: 28423670.

96. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013; 14:195–206. PMID: 23954158.

97. Fardini Y, Wang X, Témoin S, et al. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol. 2011; 82:1468–1480. PMID: 22040113.

98. Sears CL, Islam S, Saha A, et al. Association of enterotoxigenic Bacteroides fragilis infection with inflammatory diarrhea. Clin Infect Dis. 2008; 47:797–803. PMID: 18680416.

99. Toprak NU, Yagci A, Gulluoglu BM, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006; 12:782–786. PMID: 16842574.

100. Rocha ER, Smith CJ. Ferritin-like family proteins in the anaerobe Bacteroides fragilis: when an oxygen storm is coming, take your iron to the shelter. Biometals. 2013; 26:577–591. PMID: 23842847.

101. Betteken MI, Rocha ER, Smith CJ. Dps and DpsL mediate survival in vitro and in vivo during the prolonged oxidative stress response in Bacteroides fragilis. J Bacteriol. 2015; 197:3329–3338. PMID: 26260459.

102. Park Y, Choi JY, Yong D, Lee K, Kim JM. Clinical features and prognostic factors of anaerobic infections: a 7-year retrospective study. Korean J Intern Med. 2009; 24:13–18. PMID: 19270476.

103. Rhee KJ, Wu S, Wu X, et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect Immun. 2009; 77:1708–1718. PMID: 19188353.

104. Wick EC, Rabizadeh S, Albesiano E, et al. Stat3 activation in murine colitis induced by enterotoxigenic Bacteroides fragilis. Inflamm Bowel Dis. 2014; 20:821–834. PMID: 24704822.

105. Kuo WT, Lee TC, Yang HY, et al. LPS receptor subunits have antagonistic roles in epithelial apoptosis and colonic carcinogenesis. Cell Death Differ. 2015; 22:1590–1604. PMID: 25633197.

106. Kuo WT, Lee TC, Yu LC. Eritoran suppresses colon cancer by altering a functional balance in Toll-like receptors that bind lipopolysaccharide. Cancer Res. 2016; 76:4684–4695. PMID: 27328732.

107. Kuo WT, Lee TC, Yu LC. Janus-faced bacterial regulation of epithelial cell death and survival: association with colon carcinogenesis. Mol Cell Oncol. 2015; 3:e1029064. DOI: 10.1080/23723556.2015.1029064. PMID: 27308544.

108. Fukata M, Abreu MT. TLR4 signalling in the intestine in health and disease. Biochem Soc Trans. 2007; 35:1473–1478. PMID: 18031248.

109. Yu LC, Wei SC, Ni YH. Interplay between the gut microbiota and epithelial innate signaling in colitis-associated colon carcinogenesis. Cancer Res Front. 2017; 3:1–28.

110. Chen TL, Chen S, Wu HW, et al. Persistent gut barrier damage and commensal bacterial influx following eradication of Giardia infection in mice. Gut Pathog. 2013; 5:26. PMID: 23991642.

111. Kalischuk LD, Leggett F, Inglis GD. Campylobacter jejuni induces transcytosis of commensal bacteria across the intestinal epithelium through M-like cells. Gut Pathog. 2010; 2:14. PMID: 21040540.

112. Kalischuk LD, Inglis GD, Buret AG. Campylobacter jejuni induces transcellular translocation of commensal bacteria via lipid rafts. Gut Pathog. 2009; 1:2. PMID: 19338680.

113. Denizot J, Sivignon A, Barreau F, et al. Adherent-invasive Escherichia coli induce claudin-2 expression and barrier defect in CEABAC10 mice and Crohn's disease patients. Inflamm Bowel Dis. 2012; 18:294–304. PMID: 21688348.

114. Lapointe TK, O'Connor PM, Jones NL, Menard D, Buret AG. Interleukin-1 receptor phosphorylation activates Rho kinase to disrupt human gastric tight junctional claudin-4 during Helicobacter pylori infection. Cell Microbiol. 2010; 12:692–703. PMID: 20070312.

115. Fedwick JP, Lapointe TK, Meddings JB, Sherman PM, Buret AG. Helicobacter pylori activates myosin light-chain kinase to disrupt claudin-4 and claudin-5 and increase epithelial permeability. Infect Immun. 2005; 73:7844–7852. PMID: 16299274.

116. Wei SC, Yang-Yen HF, Tsao PN, et al. SHANK3 regulates intestinal barrier function through modulating ZO-1 expression through the PKCε-dependent pathway. Inflamm Bowel Dis. 2017; 23:1730–1740. PMID: 28906292.

117. Salzman AL, Eaves-Pyles T, Linn SC, Denenberg AG, Szabó C. Bacterial induction of inducible nitric oxide synthase in cultured human intestinal epithelial cells. Gastroenterology. 1998; 114:93–102. PMID: 9428223.

118. Negroni A, Colantoni E, Vitali R, et al. NOD2 induces autophagy to control AIEC bacteria infectiveness in intestinal epithelial cells. Inflamm Res. 2016; 65:803–813. PMID: 27335178.

119. Huang FC. De Novo sphingolipid synthesis is essential for Salmonella-induced autophagy and human beta-defensin 2 expression in intestinal epithelial cells. Gut Pathog. 2016; 8:5. PMID: 26893616.

120. Lu C, Chen J, Xu HG, et al. MIR106B and MIR93 prevent removal of bacteria from epithelial cells by disrupting ATG16L1-mediated autophagy. Gastroenterology. 2014; 146:188–199. PMID: 24036151.

Table 1
Bacteria Promoted Tumorigenesis in in situ Mouse CRC Models
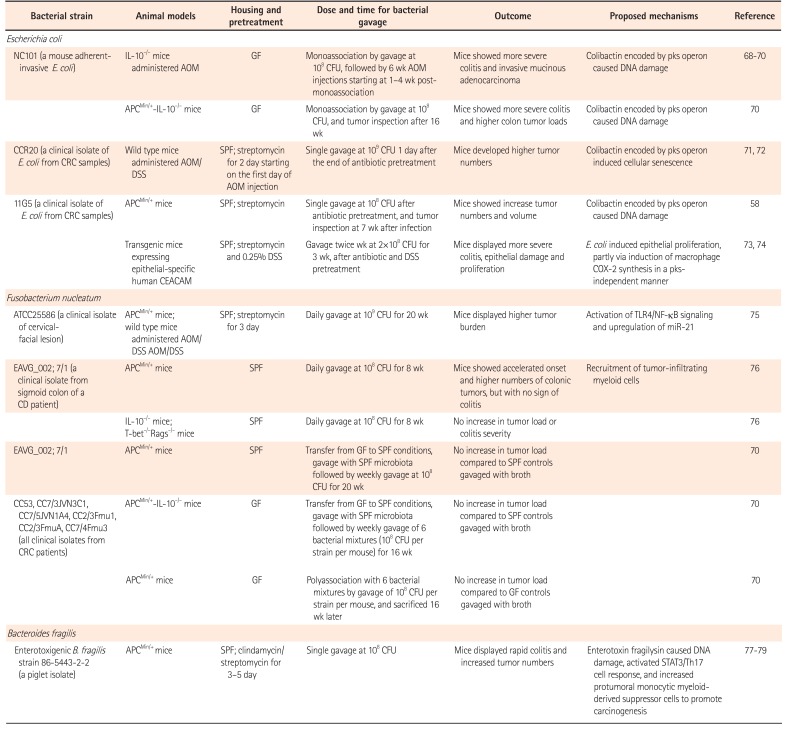
| Bacterial strain | Animal models | Housing and pretreatment | Dose and time for bacterial gavage | Outcome | Proposed mechanisms | Reference |
|---|---|---|---|---|---|---|
| Escherichia coli | ||||||
| NC101 (a mouse adherentinvasive E. coli) | IL-10−/− mice administered AOM | GF | Monoassociation by gavage at 108 CFU, followed by 6 wk AOM injections starting at 1–4 wk postmonoassociation | Mice showed more severe colitis and invasive mucinous adenocarcinoma | Colibactin encoded by pks operon caused DNA damage | 68 69 70 |
| APCMin/+-IL-10−/− mice | GF | Monoassociation by gavage at 108CFU, and tumor inspection after 16 wk | Mice showed more severe colitis and higher colon tumor loads | Colibactin encoded by pks operon caused DNA damage | 70 | |
| CCR20 (a clinical isolate of E. coli from CRC samples) | Wild type mice administered AOM/DSS | SPF; streptomycin for 2 day starting on the first day of AOM injection | Single gavage at 109 CFU 1 day after the end of antibiotic pretreatment | Mice developed higher tumor numbers | Colibactin encoded by pks operon induced cellular senescence | 71 72 |
| 11G5 (a clinical isolate of E. coli from CRC samples) | APCMin/+ mice | SPF; streptomycin | Single gavage at 108 CFU after antibiotic pretreatment, and tumor inspection at 7 wk after infection | Mice showed increase tumor numbers and volume | Colibactin encoded by pks operon caused DNA damage | 58 |
| Transgenic mice expressing epithelial-specific human CEACAM | SPF; streptomycin and 0.25% DSS | Gavage twice wk at 2×108 CFU for 3 wk, after antibiotic and DSS pretreatment | Mice displayed more severe colitis, epithelial damage and proliferation | E. coli induced epithelial proliferation, partly via induction of macrophage COX-2 synthesis in a pks-independent manner | 73 74 | |
| Fusobacterium nucleatum | ||||||
| A TCC25586 (a clinical isolate of cervical-facial lesion) | APCMin/+ mice; wild type mice administered AOM/ DSS AOM/DSS | SPF; streptomycin for 3 day | Daily gavage at 109 CFU for 20 wk | Mice displayed higher tumor burden | Activation of TLR4/NF-κB signaling and upregulation of miR-21 | 75 |
| EAVG_002; 7/1 (a clinical isolate from sigmoid colon of a CD patient) | APCMin/+ mice | SPF | Daily gavage at 108 CFU for 8 wk | Mice showed accelerated onset and higher numbers of colonic tumors, but with no sign of colitis | Recruitment of tumor-infiltrating myeloid cells | 76 |
| IL-10−/− mice; T-bet−/−Rags−/− mice | SPF | Daily gavage at 108 CFU for 8 wk | No increase in tumor load or colitis severity | 76 | ||
| EAVG_002; 7/1 | APCMin/+ mice | SPF | Transfer from GF to SPF conditions, gavage with SPF microbiota followed by weekly gavage at 108 CFU for 20 wk | No increase in tumor load compared to SPF controls gavaged with broth | 70 | |
| C53, CC7/3JVN3C1, CC7/5JVN1A4, CC2/3Fmu1, CC2/3FmuA, CC7/4Fmu3 (all clinical isolates from CRC patients) | APCMin/+-IL-10−/− mice | GF | Transfer from GF to SPF conditions, gavage with SPF microbiota followed by weekly gavage of 6 bacterial mixtures (108 CFU per strain per mouse) for 16 wk | No increase in tumor load compared to SPF controls gavaged with broth | 70 | |
| APCMin/+ mice | GF | Polyassociation with 6 bacterial mixtures by gavage of 101 CFU per strain per mouse, and sacrificed 16 wk later | No increase in tumor load compared to GF controls gavaged with broth | 70 | ||
| Bacteroides fragilis | ||||||
| Enterotoxigenic B. fragilis strain 86-5443-2-2 (a piglet isolate) | APCMin/+ mice | SPF; clindamycin/streptomycin for 3–5 day | Single gavage at 108 CFU | Mice displayed rapid colitis and increased tumor numbers | Enterotoxin fragilysin caused DNA damage, activated STAT3/Th17 cell response, and increased protumoral monocytic myeloid-derived suppressor cells to promote carcinogenesis | 77 78 79 |
CRC, colorectal cancer; IL, interleukin; AOM, azoxymethane; GF, germ free; CFU, colony-forming units; pks, polyketide synthase; APC, adenomatous polyposis coli; DSS, dextran sulfate sodium; SPF, specific pathogen-free; CEACAM, carcinoembryonic antigen-related cell adhesion molecule; COX, clyclooxygenase; TLR, Toll-like receptor; NF-κB, nuclear factor κB.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download