INTRODUCTION
Osteoporosis, which imposes a substantial medical and socioeconomic burden due to its relationship to age-related fractures [
1], manifests as impaired bone mass and microarchitecture deterioration with an elevated risk of fragility fractures [
2,
3]. Postmenopausal osteoporosis caused by a lack of estrogen is the commonest type of osteoporosis [
4]. Osteoblasts, bone-forming cells that derive from mesenchymal stem cells (MSCs), can induce bone matrix synthesis, control skeletal mineralization, and maintain bone formation through communicating with other organ systems [
5,
6]. It is of crucial importance to understand the bioenergetics of cell types such as osteoclasts, osteocytes, and adipocytes to achieve a systematic perspective on the maintenance of bone metabolism [
7]. Both environmental and genetic factors are involved in the complicated pathogenesis of primary osteoporosis. The identification of osteoporosis-related factors in the last couple of decades has contributed to an in depth understanding of its pathogenesis and the development of more therapeutic options [
8].
MicroRNAs (miRNAs) are a category of short noncoding RNAs (18 to 22 nucleotides in length) that can act as posttranscriptional regulators of gene expression [
9,
10]. Studies have elucidated the involvement of miRNAs in modulating osteoclast differentiation, bone homeostasis, and bone resorption, and also have suggested the promise of miRNA-based therapies for treating osteoporosis [
11,
12]. For instance, miR-139-3p represses the differentiation of osteoclasts (MC3T3-E1) [
13], whereas miR-98-5p promotes osteoclast differentiation [
14]. Microarray analyses and experimental validation have identified five upregulated miRNAs in the setting of healing fractures [
15], among which miR-181a-5p can decelerate the osteogenic differentiation of human bone marrow MSCs through downregulating osteoblast markers [
16]. Another study has proposed that estrogen deficiency-induced elevation of miR-181a post-menopause leads to Fas ligand deficiency, which attenuates osteoclast apoptosis and consequently drives hyperactive bone resorption and osteoporosis [
17].
A prior study has identified Runt-related transcription factor 1 (Runx1) as a downstream target of miR-181a [
18]. Runx1 is an enhancer of osteoblast differentiation and a positive regulator of osteoblast genes, ultimately contributing to bone formation [
19]. Genetic knockout of Runx1 can inhibit bone mass while boosting osteoclastogenesis and bone resorption [
20]. In light of the aforementioned findings, miR-181a-5p may target Runx1 to control the osteogenic differentiation and affect bone formation, thereby mediating the progression of osteoporosis. Therefore, we sought to verify this hypothesis in cellular and animal models of osteoporosis, by which more protective targets against osteoporosis may be unveiled.
METHODS
Ethics statement
All clinical procedures were approved by the ethics committee of the First Affiliated Hospital of Harbin Medical University (approval number: 2021003), and all participants provided written informed consent. All animal experiments were executed following the Guidelines for the Care and Use of Laboratory Animals and were approved by the Animal Care Ethics Committee of the First Affiliated Hospital of Harbin Medical University.
Clinical sample collection
Fasting venous blood samples (5 mL per individual) were collected from 56 patients who were diagnosed with osteoporosis and 48 healthy individuals who received physical examinations at the First Affiliated Hospital of Harbin Medical University. The fasting venous blood samples were placed in blood collection tubes and then centrifuged to obtain the upper serum, followed by storage at −80°C. The baseline clinical characteristics of the patients with osteoporosis and the healthy participants are displayed in
Supplemental Table S1. There were no significant differences in sex, age, weight, height, and body mass index between the patients with osteoporosis and the healthy participants.
The inclusion criteria were as follows: (1) having been diagnosed with osteoporosis by dual-energy X-ray absorptiometry (osteoporosis: T-score ≤−2.5; osteopenia: −2.5< T-score <−1.0; normal: T-score ≥−1.0); (2) not receiving anti-osteoporosis treatment before sample collection; (3) having complete clinical data and imaging data; (4) not having any psychiatric disease, with ideal cooperation; and (5) voluntarily signing the informed consent form.
The following exclusion criteria were applied: (1) being unable to cooperate due to mental and cognitive impairment; (2) having received treatment for osteopenia or osteoporosis; (3) having malignant tumors, coagulation dysfunction, allergies, liver or kidney failure, hematological diseases, diabetes, and malabsorption syndromes affecting bone metabolism; and (4) having acute-stage diseases.
Cell culture
Pre-osteoblast MC3T3-E1 cells (from the cell bank of the Chinese Academy of Sciences [Shanghai, China]) were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA). Osteoblastic differentiation of MC3T3-E1 cells was induced by supplementing a mixture of 10% FBS, dexamethasone (10−8 mol/L), L-ascorbic acid (10 mM), and β-glycerophosphate (10 mM) (Takara Bio, Shiga, Japan) every 2 days.
Cell transfection and grouping
The cells were grouped as follows: blank group, mimic-negative control (NC) group, inhibitor-NC group, shRNA-NC (sh-NC) group, miR-181a-5p-mimic group, miR-181a-5p-inhibitor group, sh-Runx1 group, miR-181a-5p-inhibitor+sh-NC group, miR-181a-5p-inhibitor+sh-Runx1 group, miR-181a-5p mimic+sh-NC group, mimic-NC+sh-allograft inflammatory factor-1 (AIF-1) group, miR-181a-5p mimic+sh-AIF-1 group, and mimic-NC+sh-NC group. The miR-181a-5p-mimic, miR-181a-5p-inhibitor, sh-Runx1, sh-AIF-1, and NCs were purchased from GeneChem (Shanghai, China). The transfection dose of the mimic was 50 nM, the transfection dose of the inhibitor was 100 nM, and the plasmid was transfected at a final concentration of 2 μg. Cells were transfected using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, and subsequent experiments were carried out 48 hours post-transfection.
Quantitative real-time polymerase chain reaction
The total RNA was extracted using the TRIzol reagent (Invitrogen), and reverse transcription was conducted with a reverse transcription kit (TaKaRa, Tokyo, Japan) referring to the kit’s requirements. Gene expression was detected using a LightCycler 480 fluorescence quantitative polymerase chain reaction (PCR) instrument (Roche, Indianapolis, IN, USA), and the reaction conditions were implemented based upon the instructions of the fluorescence quantitative PCR kit (SYBR Green Mix, Roche Diagnostics). Quantitative PCR was performed with three replicates per reaction. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 was utilized as a loading control. Data were quantified by the 2
−ΔΔCt method. The primer sequences for genes are detailed in
Supplemental Table S2.
Western blot assay
MC3T3-E1 cells were lysed with radio-immunoprecipitation assay lysis buffer (Beyotime, Shanghai, China) to obtain protein samples. Subsequently, the concentration of the protein samples was measured using a bicinchoninic acid kit (Beyotime), and the corresponding volume of protein was supplemented with loading buffer (Beyotime) to denature the protein. The protein was then subjected to separation using sodium dodecyl sulfate polyacrylamide gel electrophoresis, and the gel was electro-transferred onto a polyvinylidene fluoride membrane. The membrane was rinsed with washing solution for 1 to 2 minutes and blocked with blocking solution for 60 minutes. Next, the blot was immunoblotted for 2 hours with one of the following primary antibodies: rabbit-anti human GAPDH (ab9485, 1:5,000), Runx1 (ab92336, 1:1,000), AIF-1 (#17198, 1:500, Cell Signaling Technology, Beverly, MA, USA), collagen type I (Col1A1, ab134710, 1:5,000), osteopontin (OPN, ab214050, 1:1,000), alkaline phosphatase (ALP, ab229126, 1:2,000), and osteocalcin (OCN, ab133612, 1:4,000) (all from Abcam, Cambridge, UK except for AIF-1 antibody). Next, the blot was incubated for 1 hour with a secondary antibody: horseradish peroxidase-conjugated goat-anti rabbit immunoglobulin G (IgG) (1:5,000, Beijing ComWin Biotech Co. Ltd., Beijing, China). Lastly, the membrane was assessed with an enhanced chemiluminescence solution, and activity was detected using a chemiluminescence imaging system (Bio-Rad, Hercules, CA, USA).
Dual-luciferase reporter gene assay
The binding site between miR-181a-5p and Runx1 was predicted through the bioinformatics software StarBase. According to the predicted results, wild-type and mutant-type sequences (WT-Runx1 and MUT-Runx1) of the 3’-untranslated region of the binding site were designed, synthesized, inserted into the luciferase reporter gene vector pGL3-promoter (Promega, Madison, WI, USA), and then co-transfected into HEK293T cells with the miR-181a-5p mimic (30 nM) or its NC (30 nM) (Shanghai Sixin Biotechnology Co. Ltd., Shanghai, China).
The binding site of Runx1 and AIF-1 was predicted using the JASPAR database. The WT-AIF-1 and MUT-AIF-1 sequences of the AIF-1 promoter fragment containing the predicted Runx1 binding site were cloned into the pGL3-basic reporter vector (Promega) and co-transfected into HEK293T cells (Shanghai Sixin Biotechnology) with the overexpressed Runx1 vector or its NC. The activity of both firefly and renilla luciferase was measured with a luminescence detector (Promega) in cells posttransfection. Firefly luciferase activity was standardized based on renilla luciferase activity. Relative luciferase activity was presented as a percentage reflecting the proportion of firefly luciferase activity to renilla luciferase activity.
RNA immunoprecipitation
An RNA immunoprecipitation (RIP) assay was performed to detect the binding of miR-181a-5p to Runx1. In brief, the harvested cells were lysed with RIP lysis buffer. Next, RIP wash buffer (100 μL) was used to suspend magnetic beads, and the magnetic beads were coated with argonaute-2 (Ago2) antibody (5 μg, ab186733, 1:50, Abcam) or its NC IgG, followed by incubation overnight at 4°C on a rotator. The prepared cell lysate was quickly thawed, followed by 10 minutes of centrifugation (14,000 rpm, 4°C). After that, 100 μL of the supernatant was incubated with the magnetic bead-antibody complex at 4°C overnight and centrifuged briefly, after which the centrifuge tube was placed on a magnetic rack to remove the supernatant. The complex was then supplemented with RIP wash buffer (500 μL), and the centrifuge tube was vibrated and placed on a magnetic rack to remove the supernatant. After washing each sample six times, 150 μL of proteinase K buffer was added to resuspend the magnetic bead-antibody complex, followed by 30 minutes of incubation with the complex at 55°C; the sample was placed on a magnetic rack to obtain the supernatant. The purified RNAs were evaluated by quantitative real-time polymerase chain reaction (qRT-PCR) analysis.
Chromatin immunoprecipitation
A chromatin immunoprecipitation (ChIP) assay was implemented by using a ChIP detection kit (Millipore, Billerica, MA, USA). Briefly, cellular chromatin was cultured overnight at 4°C with Runx1 antibody (ab1109443, 1:100, Abcam) for full binding. Next, the complex of protein and DNA was precipitated by 1 hour of incubation at 4°C with the protein G agarose. After centrifugation at 5,000 ×g for 1 minute to remove the supernatant, the complex of protein and DNA was eluted. Subsequently, the cross-linking was de-linked at 65°C overnight, and the DNA fragments were then subjected to purification and recycling. The fragment region of the predicted site in the AIF-1 promoter region was amplified by PCR.
Flow cytometry
MC3T3-E1 cells were trypsinized, washed with phosphate-buffered saline (PBS), and centrifuged at 1,000 rpm for 5 minutes. Next, the MC3T3-E1 cells were resuspended in PBS, and dyed with an annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (BioVision, Milpitas, CA, USA). The apoptosis rate was evaluated by a flow cytometer (BD Biosciences, San Jose, CA, USA).
Alizarin red S staining
Alizarin red S staining was conducted to identify the mineralized nodules of osteoblasts. In short, MC3T3-E1 cells at 8 days after consecutive culture were collected, and the cell culture medium was removed. Subsequently, the MC3T3-E1 cells were subjected to fixation with ethanol (95%), followed by the removal of ethanol and washing with PBS twice. The MC3T3-E1 cells were then dyed with 0.2% alizarin red S stain (2 mL) for 30 minutes, slightly washed in running water, blocked with a neutral gum, and captured under an optical microscope. The quantification and statistical analysis of the results were performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA) and GraphPad Prism 8 software (GraphPad LLC, San Diego, CA, USA), respectively.
ALP staining
Cells at 0, 4, 8, and 12 days after induction were fixed, rinsed twice with double-distilled H2O, and then stained using a BCIP/NBT ALP chromogenic kit (Beyotime), referring to the manufacturer’s instructions. Next, the cells were washed and then photomicrographed (Olympus, Tokyo, Japan). The quantification and statistical analysis of the results were performed using ImageJ software and GraphPad Prism 8 software, respectively.
ALP enzyme assay
ALP kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) was utilized to investigate the level of ALP in MC3T3-E1 cells based on the product manual.
Establishment of an ovariectomy-induced osteoporosis mouse model
Twenty-four female specific pathogen-free C57BL/6J mice (Beijing Vital River Laboratory Animal Technology Co. Ltd., Beijing, China) were reared under standard animal feeding conditions with a light-dark 12:12-hour alternation and free access to food and water. The mice were randomly assigned into the following four groups: the sham group (no ovariectomy [OVX] mice), the OVX group (OVX), antagomir-miR-181a-5p group (OVX+antagomir-miR-181a-5p), and antagomir-NC group (OVX+antagomir-NC) (n=6).
The mice were anesthetized with 2% pentobarbital sodium (45 mg/kg) for OVX or a sham operation at 8 weeks of age. At 16 weeks of age, each animal was injected with 0.2 mL of antagomir-miR-181a-5p or antagomir-NC via the tail vein at a dose of 80 mg/kg, and the mice in the control group were injected with the same volume of PBS every week, twice a week, for 4 consecutive weeks. Lastly, the mice were euthanized, and serum samples and femurs were collected.
Hematoxylin and eosin staining
Femur samples were decalcified in 10% ethylenediaminetetraacetic acid (EDTA) for 3 weeks, and then were dehydrated and embedded in paraffin. According to the product manual of the hematoxylin and eosin (H&E) staining kit (Beyotime), the prepared tissue sections (4 μm) were dewaxed with xylene, hydrated with gradient ethanol, dyed with hematoxylin for 5 minutes, differentiated with hydrochloric acid and ethanol for 30 seconds, and dyed with eosin for 2 minutes, followed by routine dehydration, permeabilization, blocking, and photographing under a microscope.
Micro-computed tomography detection
A SCANCO µCT 100 detector, with a scanning resolution of 8 µm and a scanning range of the entire femur, was utilized to analyze the cancellous bone area within 200 layers below the growth plate. The bone mineral density (BMD, mg/cm2), trabecular bone thickness (Tb.Th, mm), trabecular bone separation (Tb.Sp, mm), trabecular bone number (Tb.N, mm−1), bone volume fraction (BV/TV, %), and bone surface/bone volume (BS/ BV, mm−1) were analyzed.
Statistical analysis
Data were statistically analyzed using GraphPad Prism 6 software (GraphPad Software, San Diego, CA, USA), and all data were depicted as mean±standard deviation. Comparisons between two groups were analyzed using the t test, and comparisons among multiple groups were tested by one-way analysis of variance, followed by the Tukey multiple-comparison test. Statistical significance was set at P<0.05.
DISCUSSION
Osteoporosis is a systemic skeletal disorder associated with bone fragility and elevated fracture risk [
21]. miRNAs have emerged as regulators of bone development in multiple aspects and have shown promise as diagnostic markers for bone disorders, including osteoporosis [
22,
23]. In the context of bone formation, subsets of miRNAs exert roles in either early or late stages of osteoblast and osteoclast differentiation by targeting suppressors of pathways involving in these cells, thus mediating the progressive differentiation [
24]. Our study principally unveiled that miR-181a-5p curbed the differentiation of pre-osteoblast MC3T3-E1
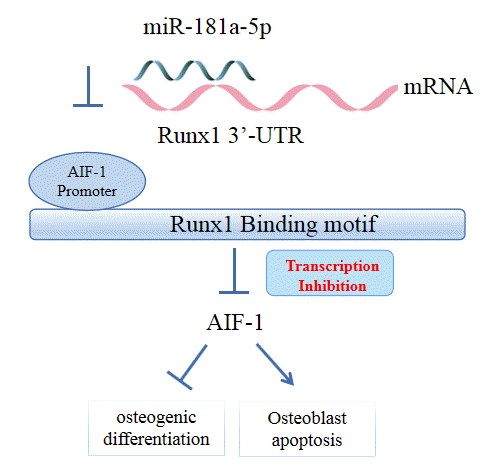
in vitro, partially through impairing Runx1-dependent inhibition of AIF-1 transcription. Of note, miR-181a-5p antagomir could alter the bone microarchitecture and augment bone formation, contributing to the prevention of postmenopausal osteoporosis.
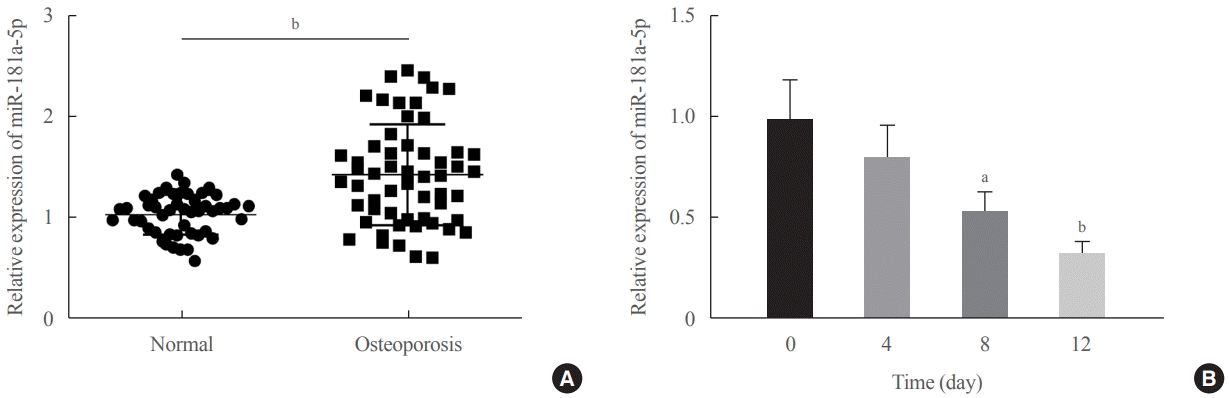
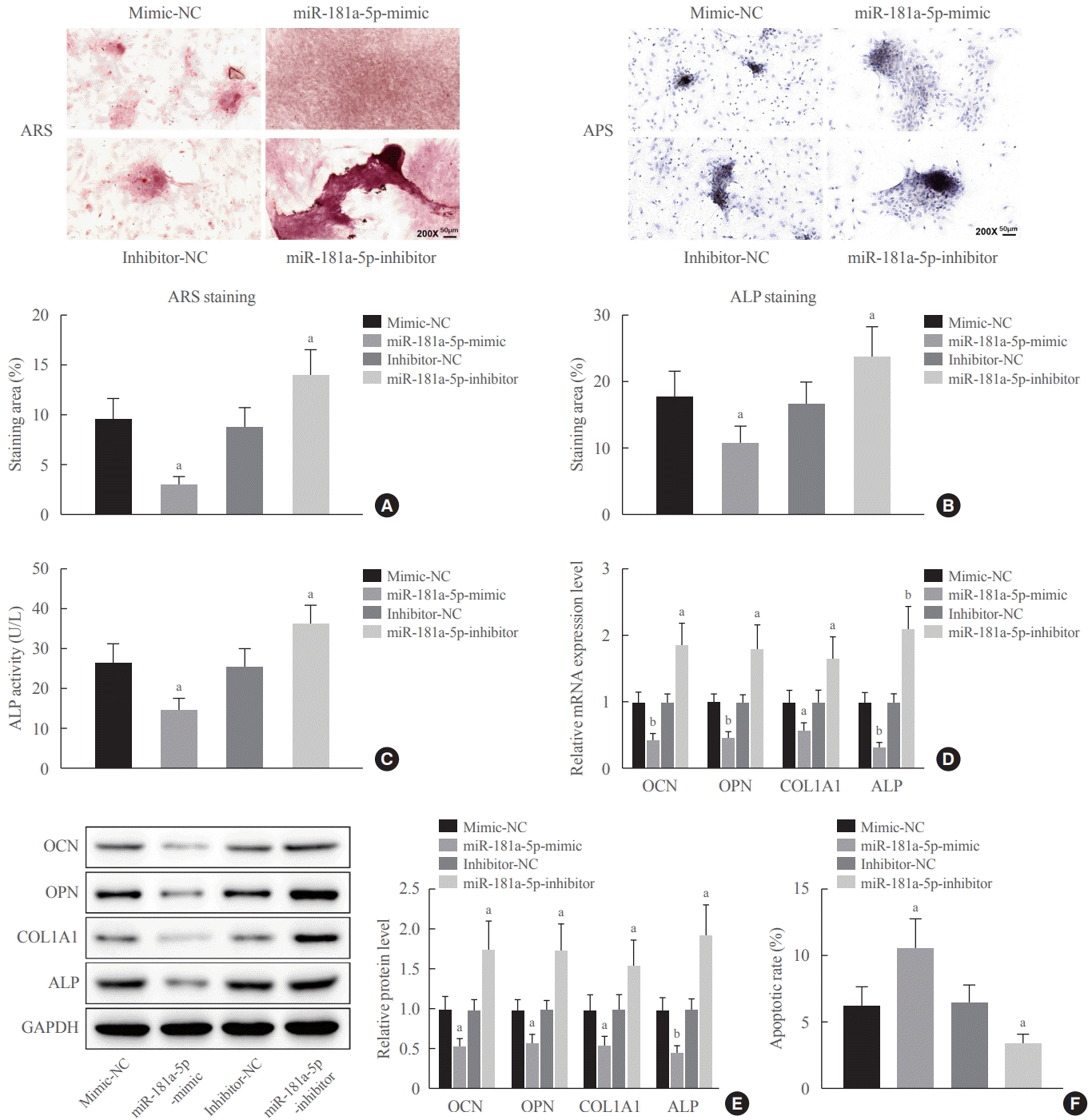
First of all, this study reported upregulated miR-181a-5p in serum samples of patients with osteoporosis, which was in line with the findings of high-throughput sequencing data in a previous study [
25]. Conversely, miR-181a-5p was reduced in a time-dependent fashion following the induction of MC3T3-E1 cell differentiation. Thus, a miR-181a-5p mimic was introduced into OIM-induced MC3T3-E1 cells, which rescued the expression of miR-181a-5p and consequently decelerated the osteoblast differentiation and matrix mineralization, accompanied by accelerated cell apoptosis. At the molecular levels, the mRNA and protein expression patterns of OCN, OPN, Col1A1, and ALP were reduced upon miR-181a-5p gain-of-function in MC3T3-E1 cells. Similarly, miR-181a-5p re-expression downregulates the levels of OPN, OCN, and Runt-related transcription factor 2 (Runx2) in human bone marrow MSCs, leading to limited osteogenic differentiation [
25]. Another study has also demonstrated that miR-181a targets an osteoblast-related transcription factor (Msx2) and negatively modulates osteoblastic differentiation in induced pluripotent stem cells [
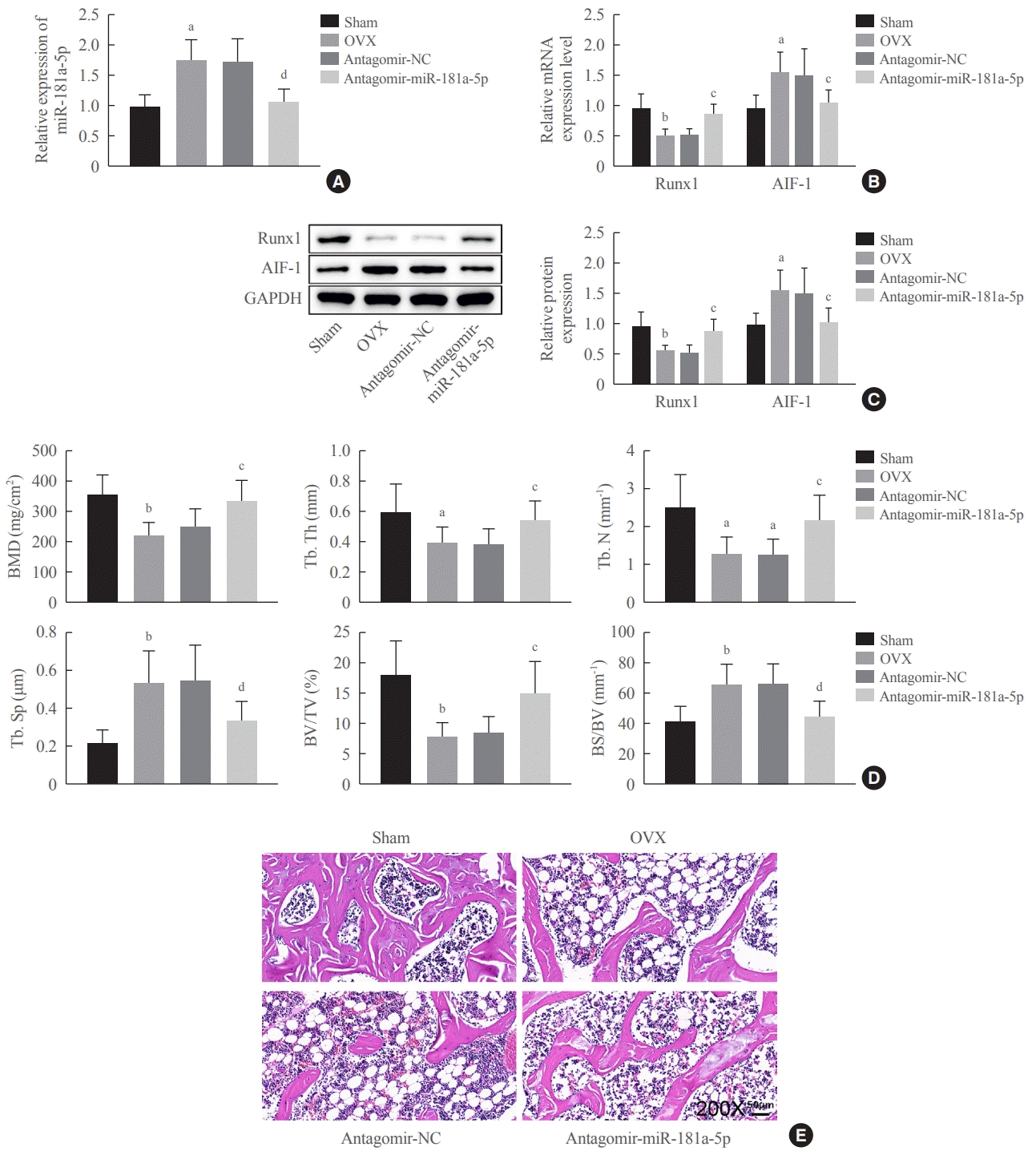
26]. The animal experiments in our study offered additional evidence, suggesting that targeting miR-181a-5p could enhance bone formation in OVX-induced mice rendered with osteoporosis, showing its promise as a therapeutic strategy against postmenopausal osteoporosis. However, the mechanisms underpinning the inhibitory role of miR-181a-5p in osteogenic differentiation remain undefined, which prompted us to decipher possible downstream targets of miR-181a-5p during this process.
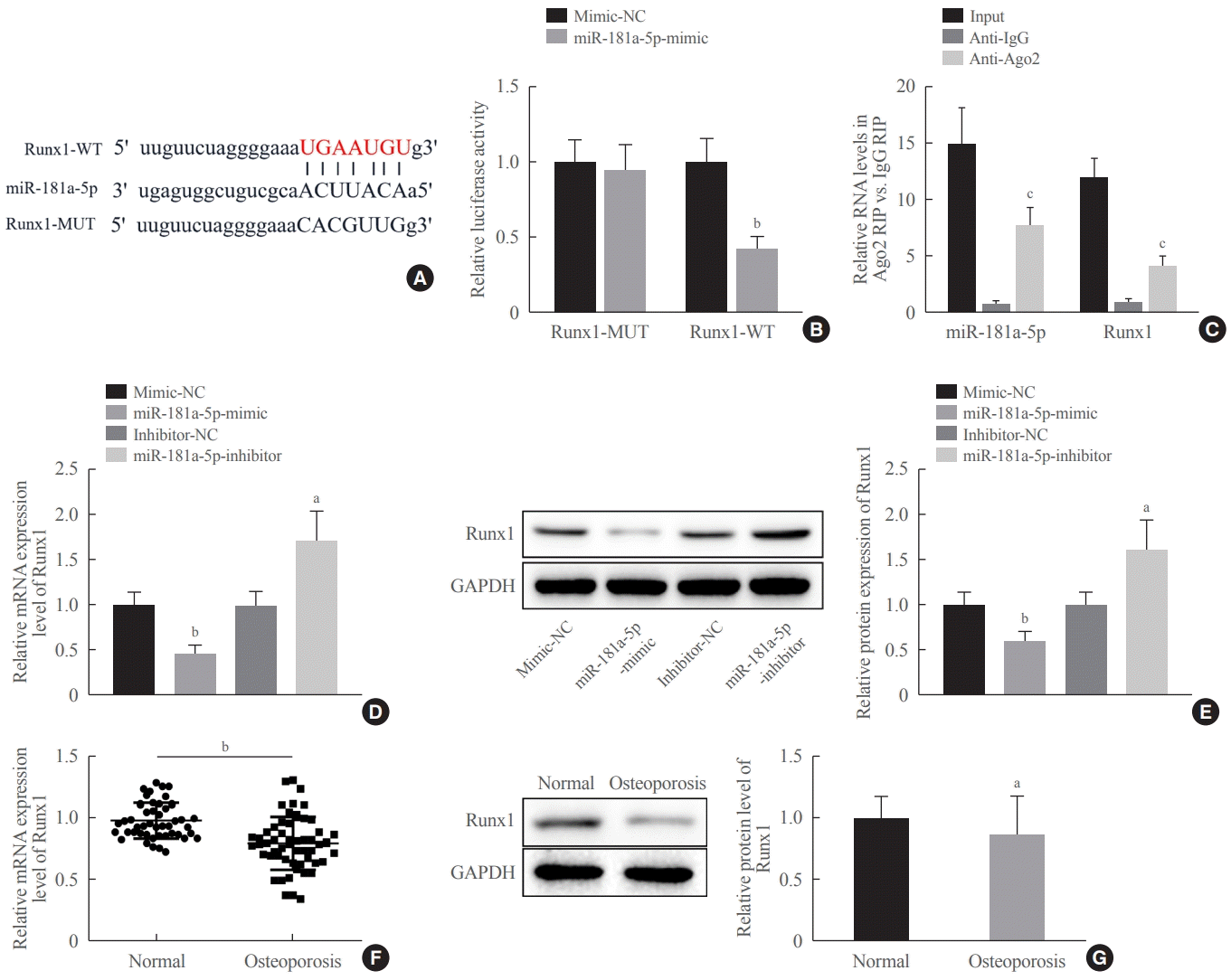
Our results from the StarBase database and dual-luciferase reporter gene assay uncovered that miR-181a-5p could directly bind to Runx1 and inversely modulated the expression of this gene. It has been proposed that Runx1 acts as a target of miR-181a based on the prediction results of TargetScan and validation results from a dual-luciferase reporter gene assay [
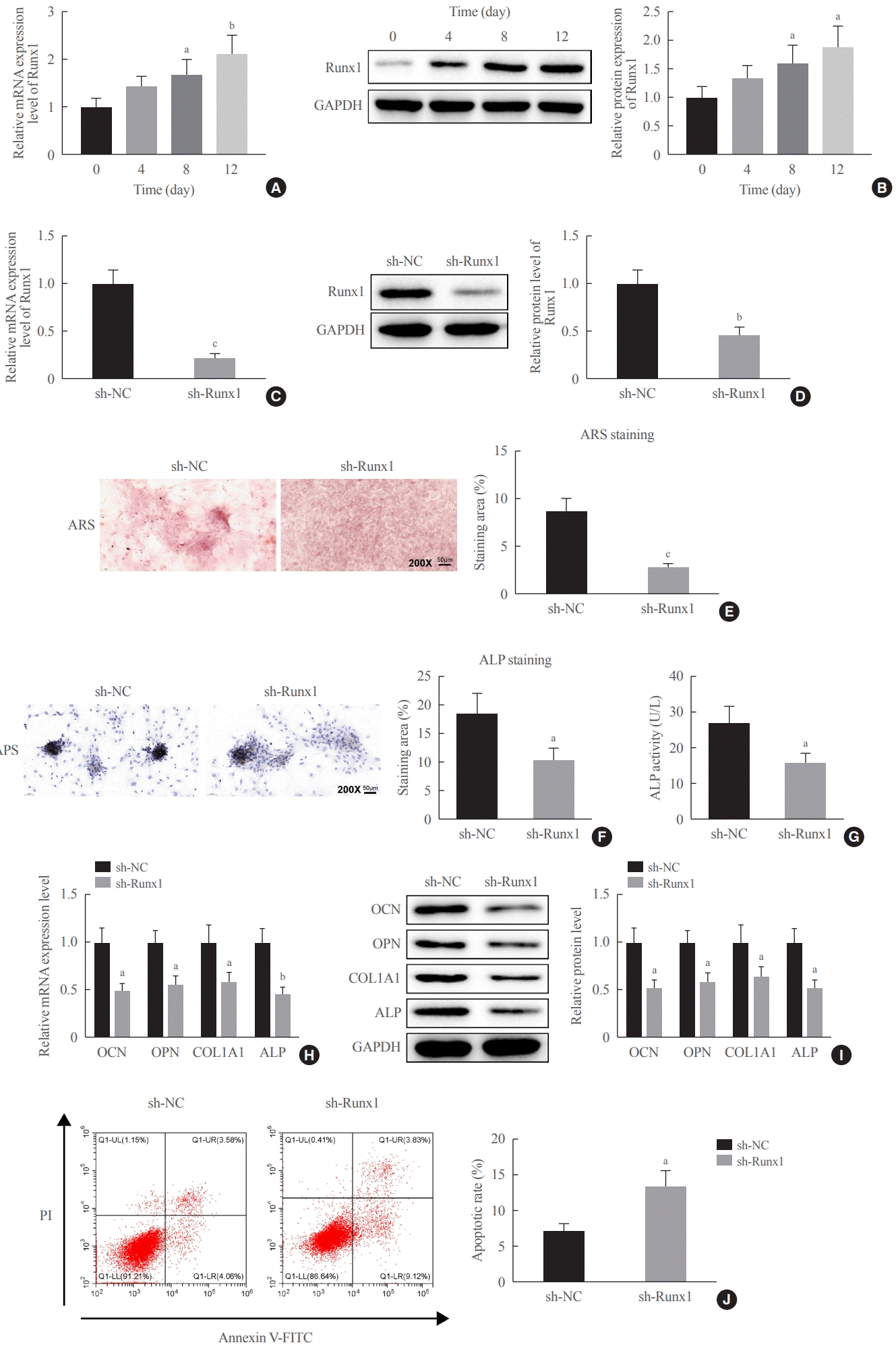
18]. Furthermore, we experimentally determined that Runx1 was poorly expressed in the serum of patients with osteoporosis and it was time-dependently elevated during osteogenic differentiation. The relationship of Runx1 to osteogenic differentiation has been widely reported. Runx1 knockdown downregulates genes and proteins pertaining to osteogenic differentiation (osterix [OSX], OCN, and OPN), and Runx1 silencing also curbs osteogenesis in bone marrow MSCs by attenuating osteogenic differentiation and mineralization [
27]. Not only can Runx1 loss in chondrocytes impede osteoblast differentiation, but its genetic deletion in mice can also downregulate osteogenic differentiation modulators (OSX, Runx2, activating transcription factor 4 [ATF4]) and osteoblast markers (OCN, OPN) in the osteoblasts, contributing to suppressed trabecular bone formation [
28]. Consistently, another study has reported that Runx1 depletion induced reductions in OCN, OSX, ATF4, and Runx2 levels in osteoblasts [
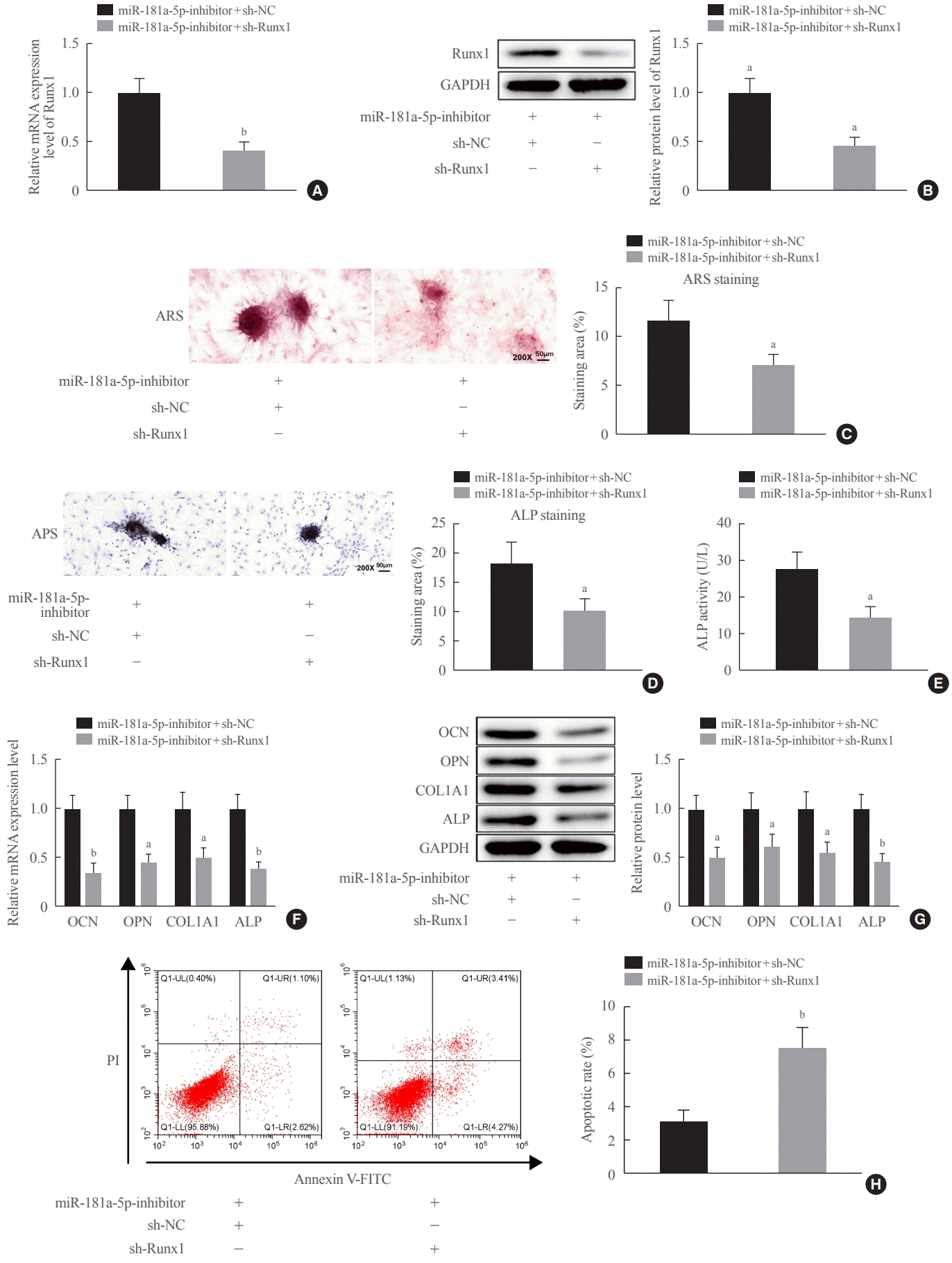
29]. This study likewise observed that shRNA-based silencing of Runx1 hindered osteogenic differentiation and matrix mineralization and facilitated the apoptosis of MC3T3-E1 cells, corresponding to reduced OCN, OPN, Col1A1, and ALP levels.
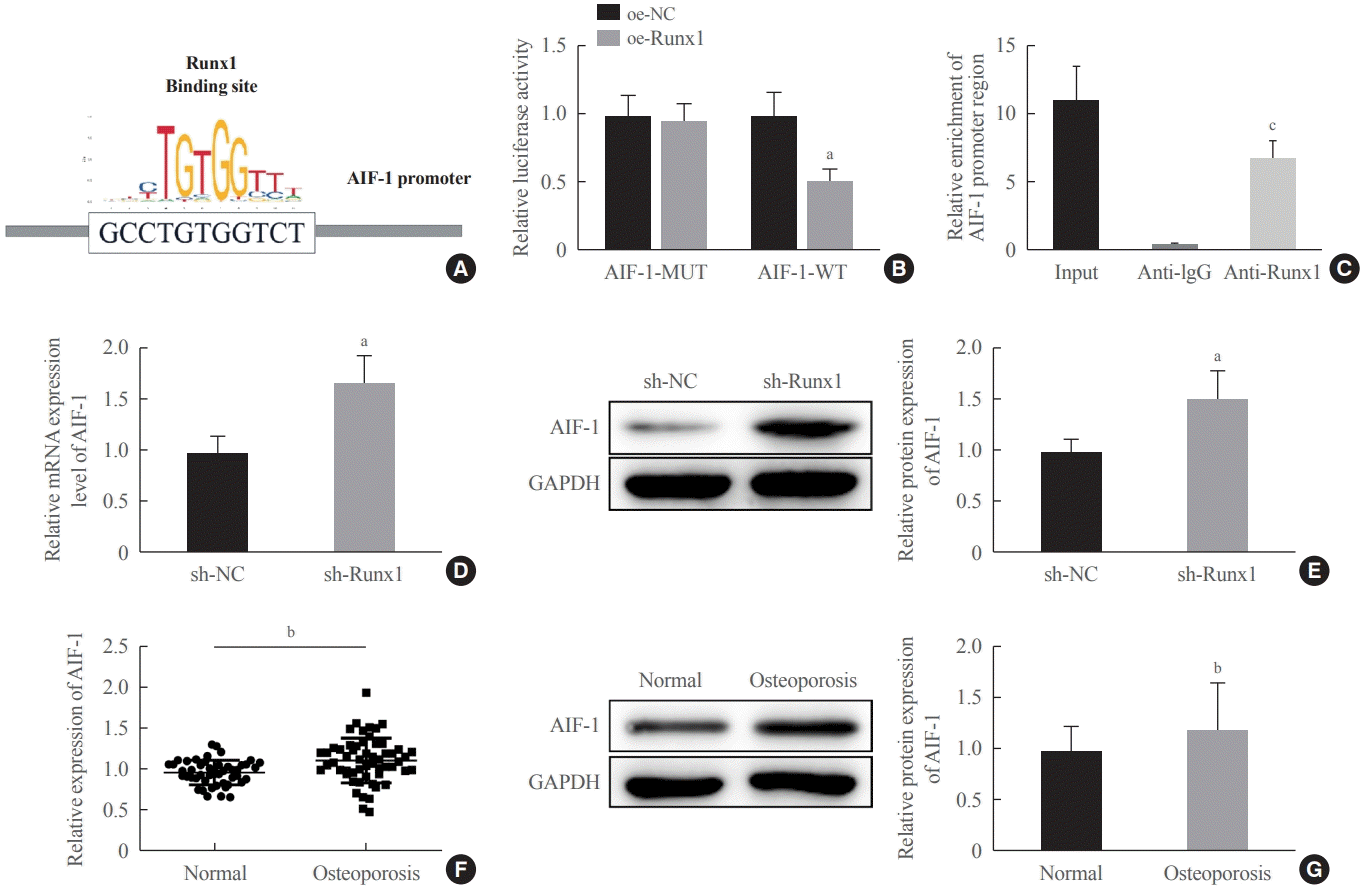
Last but not least, a mechanistic investigation by JASPAR further suggested that Runx1 could bind to the AIF-1 promoter. Thereafter, luciferase and ChIP assays validated the strong binding affinity between Runx1 and the AIF-1 promoter. Thus, Runx1 acted as a transcription factor to impair the transcription of AIF-1, which was abundantly expressed in the serum of patients with osteoporosis. AIF-1 is a 17-kDa cytoplasmic calcium-binding protein principally originating from monocytes, macrophages, and lymphocytes that is involved in chronic inflammation and potentially engaged in the pathogenesis of atherosclerosis, diabetes, and metabolic disturbances [
30]. Silencing of AIF1 has been reported to restrain the differentiation of bone marrow cells or monocytes into macrophages and that of hematopoietic stem progenitors into dendritic cells [
31,
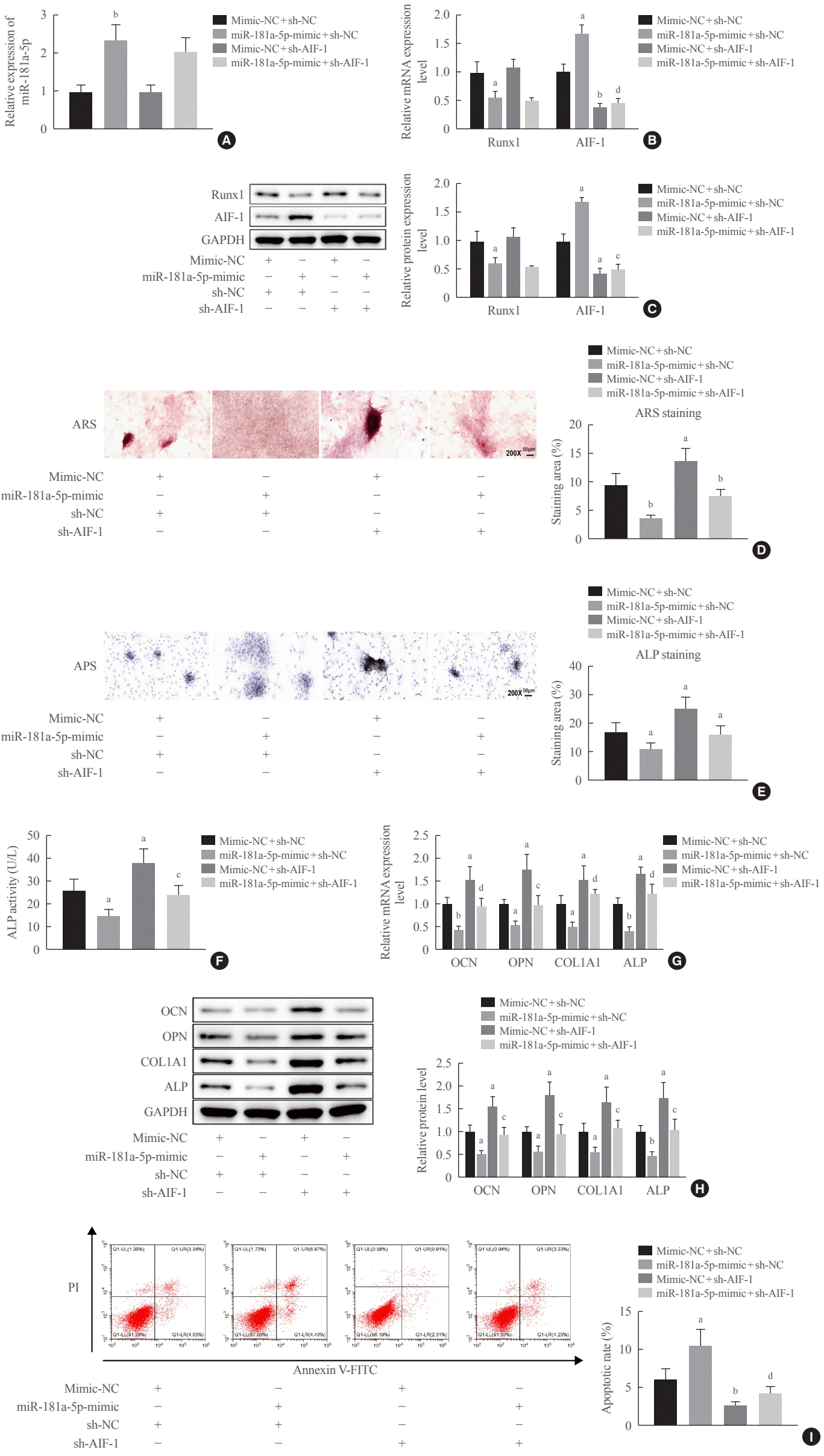
32]. In the current study, miR-181a-5p mimic elevated the expression of AIF-1, while AIF-1 knockdown caused enhanced deposition of calcified nodules and ALP activity in the presence of miR-181a-5p, indicating that miR-181a-5p might promote the transcription of AIF-1 by targeting Runx1 to repress osteogenic differentiation. Due to the lack of information relating AIF-1 to either osteogenic differentiation or osteoporosis, more studies are required to confirm the involvement of AIF-1 in the process of osteogenic differentiation.
Our study demonstrated that miR-181a-5p inhibition expedited osteogenic differentiation and bone formation partially by promoting Runx1 expression and subsequently inhibiting AIF-1 transcription. This study contributes to the development of miRNA-targeted therapeutics for osteoporosis and may broaden the current understanding of the miRNA-mRNA interactions involved in bone formation. Future research can explore other possible targets of miR-181a-5p and downstream factors of Runx1 responsible for the effects of miR-181a-5p.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download